The Challenges of Vaccine Development against Betacoronaviruses: Antibody Dependent Enhancement and Sendai Virus as a Possible Vaccine Vector
- PMID: 32921819
- PMCID: PMC7473411
- DOI: 10.1134/S0026893320060151
The Challenges of Vaccine Development against Betacoronaviruses: Antibody Dependent Enhancement and Sendai Virus as a Possible Vaccine Vector
Abstract
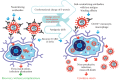
To design an effective and safe vaccine against betacoronaviruses, it is necessary to use their evolutionarily conservative antigenic determinants that will elicit the combination of strong humoral and cell-mediated immune responses. Targeting such determinants minimizes the risk of antibody-dependent enhancement of viral infection. This phenomenon was observed in animal trials of experimental vaccines against SARS-CoV-1 and MERS-CoV that were developed based on inactivated coronavirus or vector constructs expressing the spike protein (S) of the virion. The substitution and glycosylation of certain amino acids in the antigenic determinants of the S-protein, as well as its conformational changes, can lead to the same effect in a new experimental vaccine against SARS-CoV-2. Using more conservative structural and accessory viral proteins for the vaccine antigenic determinants will help to avoid this problem. This review outlines approaches for developing vaccines against the new SARS-CoV-2 coronavirus that are based on non-pathogenic viral vectors. For efficient prevention of infections caused by respiratory pathogens the ability of the vaccine to stimulate mucosal immunity in the respiratory tract is important. Such a vaccine can be developed using non-pathogenic Sendai virus vector, since it can be administered intranasally and induce a mucosal immune response that strengthens the antiviral barrier in the respiratory tract and provides reliable protection against infection.
Keywords: ADE; COVID-19; SARS-CoV-1; SARS-CoV-2; Sendai virus; antibody-dependent enhancement; conservative antigenic determinants; murine respirovirus; vaccine vector.
© The Author(s) 2020, ISSN 0026-8933, Molecular Biology, 2020, Vol. 54, No. 6, pp. 812–826. © The Author(s) 2020. This article is an open access publication.Published in Russian in Molekulyarnaya Biologiya, 2020, Vol. 54, No. 6, pp. 922–938.
Conflict of interest statement
COMPLIANCE WITH ETHICAL STANDARDSThis work did not involve humans or animals as research subjects. Conflict of interest. Authors have no conflict of interest to declare.
Figures


Similar articles
-
[The Challenges of Vaccine Development Against Betacoronaviruses: Antibody Dependent Enhancement and Sendai Virus as a Possible Vaccine Vector].Mol Biol (Mosk). 2020 Nov-Dec;54(6):922-938. doi: 10.31857/S0026898420060154. Mol Biol (Mosk). 2020. PMID: 33276356 Review. Russian.
-
The British variant of the new coronavirus-19 (Sars-Cov-2) should not create a vaccine problem.J Biol Regul Homeost Agents. 2021 Jan-Feb;35(1):1-4. doi: 10.23812/21-3-E. J Biol Regul Homeost Agents. 2021. PMID: 33377359
-
Cross-Protection against MERS-CoV by Prime-Boost Vaccination Using Viral Spike DNA and Protein.J Virol. 2020 Nov 23;94(24):e01176-20. doi: 10.1128/JVI.01176-20. Print 2020 Nov 23. J Virol. 2020. PMID: 32967955 Free PMC article.
-
Antibody Dependent Enhancement Due to Original Antigenic Sin and the Development of SARS.Front Immunol. 2020 Jun 5;11:1120. doi: 10.3389/fimmu.2020.01120. eCollection 2020. Front Immunol. 2020. PMID: 32582200 Free PMC article.
-
Insights into COVID-19 Vaccine Development Based on Immunogenic Structural Proteins of SARS-CoV-2, Host Immune Responses, and Herd Immunity.Cells. 2021 Oct 29;10(11):2949. doi: 10.3390/cells10112949. Cells. 2021. PMID: 34831172 Free PMC article. Review.
Cited by
-
The Role of Acidosis in the Pathogenesis of Severe Forms of COVID-19.Biology (Basel). 2021 Aug 31;10(9):852. doi: 10.3390/biology10090852. Biology (Basel). 2021. PMID: 34571729 Free PMC article. Review.
-
ERDRP-0519 inhibits feline coronavirus in vitro.BMC Vet Res. 2022 Jan 25;18(1):55. doi: 10.1186/s12917-022-03153-3. BMC Vet Res. 2022. PMID: 35078478 Free PMC article.
-
Theoretical Explanation for the Rarity of Antibody-Dependent Enhancement of Infection (ADE) in COVID-19.Int J Mol Sci. 2022 Sep 26;23(19):11364. doi: 10.3390/ijms231911364. Int J Mol Sci. 2022. PMID: 36232664 Free PMC article.
-
Hybrid Model of the Collapse of the Commercial Crab Paralithodes camtschaticus (Decapoda, Lithodidae) Population of the Kodiak Archipelago.Biophysics (Oxf). 2022;67(2):300-319. doi: 10.1134/S0006350922020166. Epub 2022 Jun 29. Biophysics (Oxf). 2022. PMID: 35789555 Free PMC article.
-
Efficacy of a live attenuated highly pathogenic PRRSV vaccine against a NADC30-like strain challenge: implications for ADE of PRRSV.BMC Vet Res. 2021 Jul 31;17(1):260. doi: 10.1186/s12917-021-02957-z. BMC Vet Res. 2021. PMID: 34332554 Free PMC article.
References
-
- Tirado S.M., Yoon K.J. Antibody-dependent enhancement of virus infection and disease. Viral. Immunol. 2003;16:69–86. - PubMed
-
- Jaume M., Yip M.S., Kam Y.W., Cheung C.Y., Kien F., Roberts A., Li P.H., Dutry I., Escriou N., Daeron M., Bruzzone R., Subbarao K., Peiris J.S., Nal B., Altmeyer R. SARS CoV subunit vaccine: Antibody-mediated neutralisation and enhancement. Hong Kong Med. J. 2012;18:31–36. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
