Construction of a circRNA-Related ceRNA Prognostic Regulatory Network in Breast Cancer
- PMID: 32922032
- PMCID: PMC7455596
- DOI: 10.2147/OTT.S266507
Construction of a circRNA-Related ceRNA Prognostic Regulatory Network in Breast Cancer
Abstract
Purpose: Accumulating evidence has indicated that circRNAs are closely involved in tumorigenesis and progression of human cancers. However, the molecular mechanism underlying function of circRNAs in breast cancer has not been thoroughly elucidated. Currently, we aimed to characterize the circRNA-related competing endogenous RNA (ceRNA) regulatory network in breast cancer and to construct prognostic model.
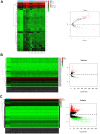
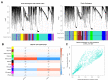
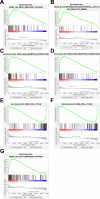
Materials and methods: First, we constructed circRNA expression profiles for paired breast cancer in a Chinese population using a human circRNA microarray. Expression profiles of circRNAs, miRNAs, and mRNAs were retrieved from our circRNA dataset, the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases. We applied the limma and edgeR packages to identify differentially expressed RNAs. Weighted gene correlation network analysis (WGCNA) was used to identify critical modules of mRNAs. Next, a ceRNA network was established based on circRNA-miRNA and miRNA-mRNA intersections. Both Cox regression analysis and ROC curve analysis were performed to generate prognostic model. Additionally, we performed Gene Set Enrichment Analysis (GSEA) on prognostic signatures.
Results: Total of 59 circRNAs, 98 miRNAs and 3966 mRNAs were identified as differentially expressed RNAs. We first identified 38 miRNA-mRNA pairs and 38 circRNA-miRNA pairs to construct the circRNA-miRNA-mRNA regulatory network and then generated a prognostic model based on 7 signatures (MMD, SLC29A4, CREB5, FOS, ANKRD29, MYOCD, and PIGR), and patients with high-risk scores presented poor prognosis. Several cancer-related pathways were enriched, including the TGF-β pathway, the focal adhesion pathway, and the JAK-STAT signaling pathway, and 20 prognostic ceRNA regulatory networks were subsequently identified.
Conclusion: In all, we screened a series of dysregulated circRNAs, miRNAs, and mRNAs, and constructed circRNA-related ceRNA network in breast cancer. Our findings may help to deepen the understanding of circRNA-related regulatory mechanisms. Moreover, we generated a prognostic model that provided new insight into postoperative management for breast cancer.
Keywords: TCGA; breast cancer; ceRNA; circRNA; mRNA; miRNA; ncRNA.
© 2020 Song et al.
Conflict of interest statement
The authors report no conflicts of interest for this work.
Figures



Similar articles
-
Integrated Analysis of Circular RNA-Associated ceRNA Network Reveals Potential circRNA Biomarkers in Human Breast Cancer.Comput Math Methods Med. 2021 Dec 20;2021:1732176. doi: 10.1155/2021/1732176. eCollection 2021. Comput Math Methods Med. 2021. Retraction in: Comput Math Methods Med. 2023 Nov 1;2023:9780596. doi: 10.1155/2023/9780596. PMID: 34966440 Free PMC article. Retracted.
-
Identification of Serum Exosome-Derived circRNA-miRNA-TF-mRNA Regulatory Network in Postmenopausal Osteoporosis Using Bioinformatics Analysis and Validation in Peripheral Blood-Derived Mononuclear Cells.Front Endocrinol (Lausanne). 2022 Jun 9;13:899503. doi: 10.3389/fendo.2022.899503. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35757392 Free PMC article.
-
Identification of Enzalutamide Resistance-Related circRNA-miRNA-mRNA Regulatory Networks in Patients with Prostate Cancer.Onco Targets Ther. 2021 Jun 21;14:3833-3848. doi: 10.2147/OTT.S309917. eCollection 2021. Onco Targets Ther. 2021. Retraction in: Onco Targets Ther. 2023 Jul 18;16:583-584. doi: 10.2147/OTT.S430430. PMID: 34188491 Free PMC article. Retracted.
-
The functional roles of competitive endogenous RNA (ceRNA) networks in apoptosis in human cancers: The circRNA/miRNA/mRNA regulatory axis and cell signaling pathways.Heliyon. 2024 Aug 31;10(21):e37089. doi: 10.1016/j.heliyon.2024.e37089. eCollection 2024 Nov 15. Heliyon. 2024. PMID: 39524849 Free PMC article. Review.
-
Systematic analysis of circRNA biomarkers for diagnosis, prognosis and therapy in colorectal cancer.Front Genet. 2022 Oct 12;13:938672. doi: 10.3389/fgene.2022.938672. eCollection 2022. Front Genet. 2022. PMID: 36313458 Free PMC article.
Cited by
-
Identification of potentially functional circular RNAs hsa_circ_0070934 and hsa_circ_0004315 as prognostic factors of hepatocellular carcinoma by integrated bioinformatics analysis.Sci Rep. 2022 Mar 23;12(1):4933. doi: 10.1038/s41598-022-08867-w. Sci Rep. 2022. PMID: 35322101 Free PMC article.
-
Construction and validation of antibody dependent cell phagocytosis related risk model in breast cancer.Sci Rep. 2025 May 29;15(1):18916. doi: 10.1038/s41598-025-03825-8. Sci Rep. 2025. PMID: 40442365 Free PMC article.
-
Characterization of Fatty Acid Metabolism in Lung Adenocarcinoma.Front Genet. 2022 Jul 14;13:905508. doi: 10.3389/fgene.2022.905508. eCollection 2022. Front Genet. 2022. PMID: 35910199 Free PMC article.
-
A novel CREB5/TOP1MT axis confers cisplatin resistance through inhibiting mitochondrial apoptosis in head and neck squamous cell carcinoma.BMC Med. 2022 Jul 1;20(1):231. doi: 10.1186/s12916-022-02409-x. BMC Med. 2022. PMID: 35773668 Free PMC article.
-
Identification of potential shared gene signatures between periodontitis and breast cancer by integrating bulk RNA-seq and scRNA-seq data.Sci Rep. 2025 Apr 2;15(1):11216. doi: 10.1038/s41598-025-95703-6. Sci Rep. 2025. PMID: 40175565 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

