Deep learning-based cell identification and disease diagnosis using spatio-temporal cellular dynamics in compact digital holographic microscopy
- PMID: 32923059
- PMCID: PMC7449709
- DOI: 10.1364/BOE.399020
Deep learning-based cell identification and disease diagnosis using spatio-temporal cellular dynamics in compact digital holographic microscopy
Abstract
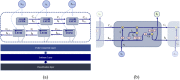
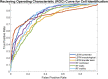
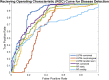
We demonstrate a successful deep learning strategy for cell identification and disease diagnosis using spatio-temporal cell information recorded by a digital holographic microscopy system. Shearing digital holographic microscopy is employed using a low-cost, compact, field-portable and 3D-printed microscopy system to record video-rate data of live biological cells with nanometer sensitivity in terms of axial membrane fluctuations, then features are extracted from the reconstructed phase profiles of segmented cells at each time instance for classification. The time-varying data of each extracted feature is input into a recurrent bi-directional long short-term memory (Bi-LSTM) network which learns to classify cells based on their time-varying behavior. Our approach is presented for cell identification between the morphologically similar cases of cow and horse red blood cells. Furthermore, the proposed deep learning strategy is demonstrated as having improved performance over conventional machine learning approaches on a clinically relevant dataset of human red blood cells from healthy individuals and those with sickle cell disease. The results are presented at both the cell and patient levels. To the best of our knowledge, this is the first report of deep learning for spatio-temporal-based cell identification and disease detection using a digital holographic microscopy system.
© 2020 Optical Society of America under the terms of the OSA Open Access Publishing Agreement.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- Schnars U., Jueptner W., Digital Holography: Digital Hologram Recording, Numerical Reconstruction, and Related Techniques (Springer, 2005).
-
- Anand A., Chhaniwal V., Patel N., Javidi B., “Automatic Identification of Malaria-Infected RBC With Digital Holographic Microscopy Using Correlation Algorithms,” IEEE Photonics J. 4(5), 1456–1464 (2012).10.1109/JPHOT.2012.2210199 - DOI
-
- Anand A., Chhaniwal V., Javidi B., “Tutorial: Common path self-referencing digital holographic microscopy,” APL Photonics 3(7), 071101 (2018).10.1063/1.5027081 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
