Multiplex detection of ctDNA mutations in plasma of colorectal cancer patients by PCR/SERS assay
- PMID: 32923312
- PMCID: PMC7484630
- DOI: 10.7150/ntno.48905
Multiplex detection of ctDNA mutations in plasma of colorectal cancer patients by PCR/SERS assay
Abstract
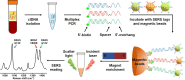
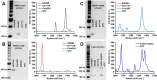
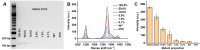
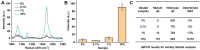
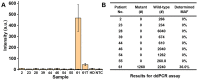
Molecular diagnostic testing of KRAS and BRAF mutations has become critical in the management of colorectal cancer (CRC) patients. Some progress has been made in liquid biopsy detection of mutations in circulating tumor DNA (ctDNA), which is a fraction of circulating cell-free DNA (cfDNA), but slow analysis for DNA sequencing methods has limited rapid diagnostics. Other methods such as quantitative PCR and more recently, droplet digital PCR (ddPCR), have limitations in multiplexed capacity and the need for expensive specialized equipment. Hence, a robust, rapid and facile strategy is needed for detecting multiple ctDNA mutations to improve the management of CRC patients. To address this significant problem, herein, we propose a new application of multiplex PCR/SERS (surface-enhanced Raman scattering) assay for the detection of ctDNA in CRC, in a fast and non-invasive manner to diagnose and stratify patients for effective treatment. Methods: To discriminate ctDNA mutations from wild-type cfDNA, allele-specific primers were designed for the amplification of three clinically important DNA point mutations in CRC including KRAS G12V, KRAS G13D and BRAF V600E. Surface-enhanced Raman scattering (SERS) nanotags were labelled with a short and specific sequence of oligonucleotide, which can hybridize with the corresponding PCR amplicons. The PCR/SERS assay was implemented by firstly amplifying the multiple mutations, followed by binding with multicolor SERS nanotags specific to each mutation, and subsequent enrichment with magnetic beads. The mutation status was evaluated using a portable Raman spectrometer where the fingerprint spectral peaks of the corresponding SERS nanotags indicate the presence of the mutant targets. The method was then applied to detect ctDNA from CRC patients under a blinded test, the results were further validated by ddPCR. Results: The PCR/SERS strategy showed high specificity and sensitivity for genotyping CRC cell lines and plasma ctDNA, where as few as 0.1% mutant alleles could be detected from a background of abundant wild-type cfDNA. The blinded test using 9 samples from advanced CRC patients by PCR/SERS assay was validated with ddPCR and showed good consistency with pathology testing results. Conclusions: With ddPCR-like sensitivity yet at the convenience of standard PCR, the proposed assay shows great potential in sensitive detection of multiple ctDNA mutations for clinical decision-making.
Keywords: CRC; PCR/SERS assay; ctDNA; ddPCR; multiplex detection.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
References
-
- Australian Institute of Health, Welfare 2019. Cancer in Australia 2019. Cancer series no.119. Cat. no. CAN 123. Canberra: AIHW.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

