Adaptive Evolution of Feline Coronavirus Genes Based on Selection Analysis
- PMID: 32923488
- PMCID: PMC7453238
- DOI: 10.1155/2020/9089768
Adaptive Evolution of Feline Coronavirus Genes Based on Selection Analysis
Abstract
Purpose: We investigated sequences of the feline coronaviruses (FCoV), which include feline enteric coronavirus (FECV) and feline infectious peritonitis virus (FIPV), from China and other countries to gain insight into the adaptive evolution of this virus.
Methods: Ascites samples from 31 cats with suspected FIP and feces samples from 8 healthy cats were screened for the presence of FCoV. Partial viral genome sequences, including parts of the nsp12-nsp14, S, N, and 7b genes, were obtained and aligned with additional sequences obtained from the GenBank database. Bayesian phylogenetic analysis was conducted, and the possibility of recombination within these sequences was assessed. Analysis of the levels of selection pressure experienced by these sequences was assessed using methods on both the PAML and Datamonkey platforms.
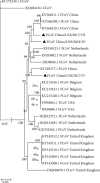
Results: Of the 31 cats investigated, two suspected FIP cats and one healthy cat tested positive for FCoV. Phylogenetic analysis showed that all of the sequences from mainland China cluster together with a few sequences from the Netherlands as a distinct clade when analyzed with FCoV sequences from other countries. Fewer than 3 recombination breakpoints were detected in the nsp12-nsp14, S, N, and 7b genes, suggesting that analyses for positive selection could be conducted. A total of 4, 12, 4, and 4 positively selected sites were detected in the nsp12-nsp14, S, N, and 7b genes, respectively, with the previously described site 245 of the S gene, which distinguishes FIPV from FECV, being a positive selection site. Conversely, 106, 168, 25, and 17 negative selection sites in the nsp12-14, S, N, and 7b genes, respectively, were identified.
Conclusion: Our study provides evidence that the FCoV genes encoding replicative, entry, and virulence proteins potentially experienced adaptive evolution. A greater number of sites in each gene experienced negative rather than positive selection, which suggests that most of the protein sequence must be conservatively maintained for virus survival. A few of the sites showing evidence of positive selection might be associated with the more severe pathology of FIPV or help these viruses survive other harmful conditions.
Copyright © 2020 Hongyue Xia et al.
Conflict of interest statement
The authors declare that they have no conflict of interests.
Figures
Similar articles
-
Mutation analysis of the spike protein in Italian feline infectious peritonitis virus and feline enteric coronavirus sequences.Res Vet Sci. 2021 Mar;135:15-19. doi: 10.1016/j.rvsc.2020.12.023. Epub 2021 Jan 2. Res Vet Sci. 2021. PMID: 33418186
-
Mutations of 3c and spike protein genes correlate with the occurrence of feline infectious peritonitis.Vet Microbiol. 2014 Oct 10;173(3-4):177-88. doi: 10.1016/j.vetmic.2014.07.020. Epub 2014 Aug 4. Vet Microbiol. 2014. PMID: 25150756 Free PMC article.
-
Circulation and genetic diversity of Feline coronavirus type I and II from clinically healthy and FIP-suspected cats in China.Transbound Emerg Dis. 2019 Mar;66(2):763-775. doi: 10.1111/tbed.13081. Epub 2018 Dec 5. Transbound Emerg Dis. 2019. PMID: 30468573 Free PMC article.
-
Feline infectious peritonitis: still an enigma?Vet Pathol. 2014 Mar;51(2):505-26. doi: 10.1177/0300985814522077. Vet Pathol. 2014. PMID: 24569616 Review.
-
Feline Coronaviruses: Pathogenesis of Feline Infectious Peritonitis.Adv Virus Res. 2016;96:193-218. doi: 10.1016/bs.aivir.2016.08.002. Epub 2016 Aug 31. Adv Virus Res. 2016. PMID: 27712624 Free PMC article. Review.
Cited by
-
Detection of Feline Coronavirus Variants in Cats without Feline Infectious Peritonitis.Viruses. 2022 Jul 29;14(8):1671. doi: 10.3390/v14081671. Viruses. 2022. PMID: 36016293 Free PMC article.
-
A review of feline infectious peritonitis virus infection.Vet World. 2024 Nov;17(11):2417-2432. doi: 10.14202/vetworld.2024.2417-2432. Epub 2024 Nov 5. Vet World. 2024. PMID: 39829669 Free PMC article. Review.
-
Natural selection differences detected in key protein domains between non-pathogenic and pathogenic Feline Coronavirus phenotypes.bioRxiv [Preprint]. 2023 Jan 11:2023.01.11.523607. doi: 10.1101/2023.01.11.523607. bioRxiv. 2023. Update in: Virus Evol. 2023 Mar 15;9(1):vead019. doi: 10.1093/ve/vead019. PMID: 36712007 Free PMC article. Updated. Preprint.
-
Epidemiology and Comparative Analyses of the S Gene on Feline Coronavirus in Central China.Pathogens. 2022 Apr 12;11(4):460. doi: 10.3390/pathogens11040460. Pathogens. 2022. PMID: 35456135 Free PMC article.
-
Feline Infectious Peritonitis: European Advisory Board on Cat Diseases Guidelines.Viruses. 2023 Aug 31;15(9):1847. doi: 10.3390/v15091847. Viruses. 2023. PMID: 37766254 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

