Hamiltonian Monte Carlo sampling to estimate past population dynamics using the skygrid coalescent model in a Bayesian phylogenetics framework
- PMID: 32923688
- PMCID: PMC7463299
- DOI: 10.12688/wellcomeopenres.15770.1
Hamiltonian Monte Carlo sampling to estimate past population dynamics using the skygrid coalescent model in a Bayesian phylogenetics framework
Abstract
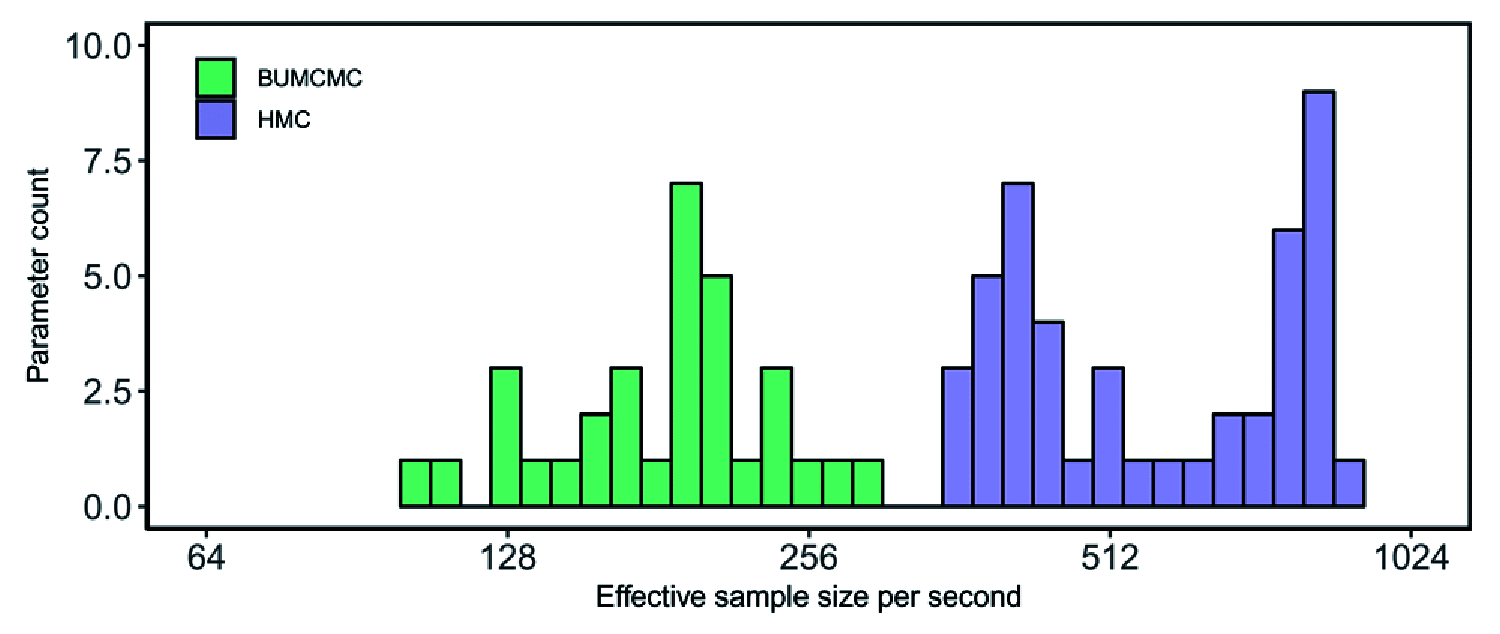
Nonparametric coalescent-based models are often employed to infer past population dynamics over time. Several of these models, such as the skyride and skygrid models, are equipped with a block-updating Markov chain Monte Carlo sampling scheme to efficiently estimate model parameters. The advent of powerful computational hardware along with the use of high-performance libraries for statistical phylogenetics has, however, made the development of alternative estimation methods feasible. We here present the implementation and performance assessment of a Hamiltonian Monte Carlo gradient-based sampler to infer the parameters of the skygrid model. The skygrid is a popular and flexible coalescent-based model for estimating population dynamics over time and is available in BEAST 1.10.5, a widely-used software package for Bayesian pylogenetic and phylodynamic analysis. Taking into account the increased computational cost of gradient evaluation, we report substantial increases in effective sample size per time unit compared to the established block-updating sampler. We expect gradient-based samplers to assume an increasingly important role for different classes of parameters typically estimated in Bayesian phylogenetic and phylodynamic analyses.
Keywords: BEAGLE; BEAST; Bayesian skygrid; Hamiltonian Monte Carlo; Markov chain Monte Carlo; pathogen phylodynamics; phylogenetics.
Copyright: © 2020 Baele G et al.
Conflict of interest statement
No competing interests were disclosed.
Figures




References
-
- Kingman JFC: On the genealogy of large populations. J Appl Probab. 1982;19(A):27–43. 10.2307/3213548 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

