Next-Generation Sequencing of Advanced GI Tumors Reveals Individual Treatment Options
- PMID: 32923905
- PMCID: PMC7446530
- DOI: 10.1200/PO.19.00359
Next-Generation Sequencing of Advanced GI Tumors Reveals Individual Treatment Options
Abstract
Purpose: Precision oncology connects highly complex diagnostic procedures with patient histories to identify individualized treatment options in interdisciplinary molecular tumor boards (MTBs). Detailed data on MTB-guided treatments and outcome with a focus on advanced GI cancers have not been reported yet.
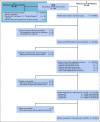
Patients and methods: Next-generation sequencing of tumor and normal tissue pairs was performed between April 2016 and February 2018. After identification of relevant molecular alterations, available clinical studies or in-label, off-label, or matched experimental treatment options were recommended. Follow-up data and a response assessment that was based on radiologic imaging were recorded.
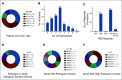
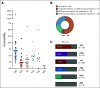
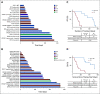
Results: Ninety-six patients were presented to the MTB of Tuebingen University Hospital. Sixteen (17%) showed "pathogenic" or "likely pathogenic" germline variants. Recommendations on the basis of molecular alterations or tumor mutational burden were given for 41 patients (43%). Twenty-five received the suggested drug, and 20 were evaluable for best response assessment. Three patients (15%) reached a partial response (PR), and 6 (30%), stable disease (SD), whereas 11 (55%) had tumor progression (progressive disease). Median progression-free survival (PFS) for all treated and evaluable patients was 2.8 months (range, 1.0-9.0 months), and median overall survival (OS) of all treated patients was 5.2 months (range, 0.1 months to not reached). Patients with SD for ≥ 3 months or PR compared with progressive disease showed both a statistically significant longer median PFS (7.8 months [95% CI, 4.2 to 11.4 months] v 2.2 months [95% CI, 1.5 to 2.8 months], P < .0001) and median OS (18.0 months [95% CI, 10.4 to 25.6 months] v 3.8 months [95% CI, 2.3 to 5.4 months], P < .0001).
Conclusion: Next-generation sequencing diagnostics of advanced GI cancers identified a substantial number of pathogenic or likely pathogenic germline variants and unique individual treatment options. Patients with PR or SD in the course of MTB-recommended treatments seemed to benefit with respect to PFS and OS.
© 2020 by American Society of Clinical Oncology.
Conflict of interest statement
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center. Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments). Michael BitzerConsulting or Advisory Role: Bristol-Myers Squibb, Bayer Vital GmbH, EISAI, Ipsen, Lilly Travel, Accommodations, Expenses: Ipsen, CelgeneSaskia BiskupEmployment: CeGaT Stock and Other Ownership Interests: CeGaTMartin SchulzeEmployment: Praxis für Humangenetik TuebingenFranz HilkeHonoraria: Agilent Technologies Research Funding: Novartis (Inst) Travel, Accommodations, Expenses: Agilent TechnologiesKonstantin NikolaouHonoraria: Siemens Healthineers, Bayer Schering Pharma Consulting or Advisory Role: Siemens Healthineers Speakers' Bureau: Siemens Healthineers Research Funding: Siemens Healthineers (Inst), Bayer Schering Pharma (Inst) Travel, Accommodations, Expenses: Siemens Healthineers, Bayer Schering PharmaChristopher SchroederResearch Funding: Illumina (Inst), Novartis (Inst)Olaf RiessHonoraria: AstraZeneca, Takeda/Shire Consulting or Advisory Role: Illumina Research Funding: Illumina (Inst)Falko FendConsulting or Advisory Role: Roche, EUSA PharmaDaniel ZipsResearch Funding: Elekta (Inst), Siemens (Inst), Sennewald (Inst) Travel, Accommodations, Expenses: Elekta (Inst)Martina HinterleitnerTravel, Accommodations, Expenses: Novartis/IpsenLars ZenderLeadership: HeparegeniX GmBH Consulting or Advisory Role: Boehringer Ingelheim Research Funding: HeparegeniX GmBH Patents, Royalties, Other Intellectual Property: Patent on MKK4 Inhibition for the treatment of acute and chronic liver diseases Travel, Accommodations, Expenses: IpsenGhazaleh TabatabaiHonoraria: AbbVie, Bayer, Medac, Novocure (Inst) Consulting or Advisory Role: AbbVie, Bayer Travel, Accommodations, Expenses: Novocure (Inst)Nisar P. MalekHonoraria: Spring Bank Travel, Accommodations, Expenses: Falk Foundation No other potential conflicts of interest were reported.
Figures
References
-
- Hoefflin R, Geißler A-L, Fritsch R, et al. Personalized clinical decision making through implementation of a molecular tumor board: A German single-center experience. JCO Precision Oncol. 2018 10.1200/PO.18.00105. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

