The Structure and Function of DNA G-Quadruplexes
- PMID: 32923997
- PMCID: PMC7472594
- DOI: 10.1016/j.trechm.2019.07.002
The Structure and Function of DNA G-Quadruplexes
Abstract
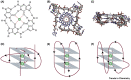
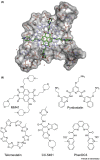
Guanine-rich DNA sequences can fold into four-stranded, noncanonical secondary structures called G-quadruplexes (G4s). G4s were initially considered a structural curiosity, but recent evidence suggests their involvement in key genome functions such as transcription, replication, genome stability, and epigenetic regulation, together with numerous connections to cancer biology. Collectively, these advances have stimulated research probing G4 mechanisms and consequent opportunities for therapeutic intervention. Here, we provide a perspective on the structure and function of G4s with an emphasis on key molecules and methodological advances that enable the study of G4 structures in human cells. We also critically examine recent mechanistic insights into G4 biology and protein interaction partners and highlight opportunities for drug discovery.
Keywords: DNA; G-quadruplex; G4; drug discovery; nucleic acids; secondary structure.
© 2019 The Author(s).
Figures


References
-
- Watson J.D., Crick F.H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. - PubMed
-
- Bang I. Untersuchungen über die Guanylsäure. Biochem. Z. 1910;26:293–311. (in German)
-
- Zimmerman S.B. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J. Mol. Biol. 1975;92:181–192. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
