Ancestral state reconstruction of metabolic pathways across pangenome ensembles
- PMID: 32924924
- PMCID: PMC7725326
- DOI: 10.1099/mgen.0.000429
Ancestral state reconstruction of metabolic pathways across pangenome ensembles
Abstract
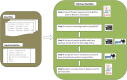
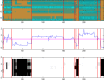
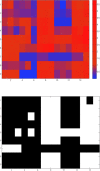
As genome sequencing efforts are unveiling the genetic diversity of the biosphere with an unprecedented speed, there is a need to accurately describe the structural and functional properties of groups of extant species whose genomes have been sequenced, as well as their inferred ancestors, at any given taxonomic level of their phylogeny. Elaborate approaches for the reconstruction of ancestral states at the sequence level have been developed, subsequently augmented by methods based on gene content. While these approaches of sequence or gene-content reconstruction have been successfully deployed, there has been less progress on the explicit inference of functional properties of ancestral genomes, in terms of metabolic pathways and other cellular processes. Herein, we describe PathTrace, an efficient algorithm for parsimony-based reconstructions of the evolutionary history of individual metabolic pathways, pivotal representations of key functional modules of cellular function. The algorithm is implemented as a five-step process through which pathways are represented as fuzzy vectors, where each enzyme is associated with a taxonomic conservation value derived from the phylogenetic profile of its protein sequence. The method is evaluated with a selected benchmark set of pathways against collections of genome sequences from key data resources. By deploying a pangenome-driven approach for pathway sets, we demonstrate that the inferred patterns are largely insensitive to noise, as opposed to gene-content reconstruction methods. In addition, the resulting reconstructions are closely correlated with the evolutionary distance of the taxa under study, suggesting that a diligent selection of target pangenomes is essential for maintaining cohesiveness of the method and consistency of the inference, serving as an internal control for an arbitrary selection of queries. The PathTrace method is a first step towards the large-scale analysis of metabolic pathway evolution and our deeper understanding of functional relationships reflected in emerging pangenome collections.
Keywords: ancestral reconstruction; comparative genomics; metabolic pathways; parsimony method; phylogenetic profiling.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
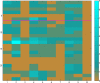






References
-
- Omland KE. The assumptions and challenges of ancestral state reconstructions. Syst Biol. 1999;48:604–611. doi: 10.1080/106351599260175. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

