Identification of Target Genes Related to Sulfasalazine in Triple-Negative Breast Cancer Through Network Pharmacology
- PMID: 32925871
- PMCID: PMC7513616
- DOI: 10.12659/MSM.926550
Identification of Target Genes Related to Sulfasalazine in Triple-Negative Breast Cancer Through Network Pharmacology
Abstract
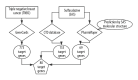
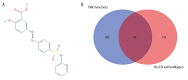
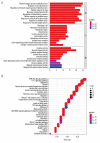
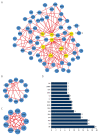
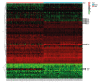
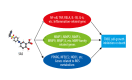
BACKGROUND The anti-inflammatory drug sulfasalazine (SAS) has been confirmed to inhibit the growth of triple-negative breast cancer (TNBC), but the mechanism is not clear. The aim of this study was to use network pharmacology to find relevant pathways of SAS in TNBC patients. MATERIAL AND METHODS Through screening of the GeneCards, CTD, and ParmMapper databases, potential genes related to SAS and TNBC were identified. In addition, gene ontology and Kyoto Encyclopedia of Genes and Genomes analyses were performed using the R programming language. Protein-protein interaction networks were constructed using Cytoscape. The Kaplan-Meier plotter screened genes related to TNBC prognosis. TNBC patient gene expression profiles and clinical data were downloaded from The Cancer Genome Atlas database. A heatmap was generated using the R programming language that presents the expression of potential target genes in patients with TNBC. RESULTS Eighty potential target genes were identified through multiple databases. The bioinformatical analyses predicted the interrelationships, potential pathways, and molecular functions of the genes from multiple aspects, which are associated with physiological processes such as the inflammatory response, metabolism of reactive oxygen species (ROS), and regulation of proteins in the matrix metalloproteinase (MMP) family. Survival analysis showed that 12 genes were correlated with TNBC prognosis. Heatmapping showed that genes such as those encoding members of the MMP family were differentially expressed in TNBC tissues and normal tissues. CONCLUSIONS Our analysis revealed that the main reasons for the inhibitory effect of SAS on TNBC cells may be inhibition of the inflammatory response and MMP family members and activation of ROS.
Conflict of interest statement
None.
Figures

Similar articles
-
Identification of differentially expressed genes between triple and non-triple-negative breast cancer using bioinformatics analysis.Breast Cancer. 2019 Nov;26(6):784-791. doi: 10.1007/s12282-019-00988-x. Epub 2019 Jun 13. Breast Cancer. 2019. PMID: 31197620
-
Integrated network analysis and machine learning approach for the identification of key genes of triple-negative breast cancer.J Cell Biochem. 2019 Apr;120(4):6154-6167. doi: 10.1002/jcb.27903. Epub 2018 Oct 9. J Cell Biochem. 2019. PMID: 30302816
-
Role and molecular mechanisms of HuangQiSiJunZi decoction for treating triple-negative breast cancer as explored via network pharmacology and bioinformatics analyses.BMC Cancer. 2024 Sep 30;24(1):1217. doi: 10.1186/s12885-024-12957-5. BMC Cancer. 2024. PMID: 39350059 Free PMC article.
-
Integrated analysis of differentially expressed genes and pathways in triple‑negative breast cancer.Mol Med Rep. 2017 Mar;15(3):1087-1094. doi: 10.3892/mmr.2017.6101. Epub 2017 Jan 4. Mol Med Rep. 2017. PMID: 28075450 Free PMC article.
-
Identification of Key Prognostic Genes of Triple Negative Breast Cancer by LASSO-Based Machine Learning and Bioinformatics Analysis.Genes (Basel). 2022 May 18;13(5):902. doi: 10.3390/genes13050902. Genes (Basel). 2022. PMID: 35627287 Free PMC article.
Cited by
-
Oral Conventional Synthetic Disease-Modifying Antirheumatic Drugs with Antineoplastic Potential: a Review.Dermatol Ther (Heidelb). 2022 Apr;12(4):835-860. doi: 10.1007/s13555-022-00713-1. Epub 2022 Apr 5. Dermatol Ther (Heidelb). 2022. PMID: 35381976 Free PMC article. Review.
-
Curcumin and its nano-formulations: Defining triple-negative breast cancer targets through network pharmacology, molecular docking, and experimental verification.Front Pharmacol. 2022 Aug 8;13:920514. doi: 10.3389/fphar.2022.920514. eCollection 2022. Front Pharmacol. 2022. PMID: 36003508 Free PMC article.
-
Cystine/cysteine metabolism regulates the progression and response to treatment of triple‑negative breast cancer (Review).Oncol Lett. 2024 Aug 30;28(5):521. doi: 10.3892/ol.2024.14654. eCollection 2024 Nov. Oncol Lett. 2024. PMID: 39268159 Free PMC article. Review.
-
Programmed cell death, redox imbalance, and cancer therapeutics.Apoptosis. 2021 Aug;26(7-8):385-414. doi: 10.1007/s10495-021-01682-0. Epub 2021 Jul 8. Apoptosis. 2021. PMID: 34236569 Review.
-
Network Pharmacology Prediction and Molecular Docking-Based Strategy to Explore the Potential Mechanism of Gualou Xiebai Banxia Decoction against Myocardial Infarction.Genes (Basel). 2024 Mar 22;15(4):392. doi: 10.3390/genes15040392. Genes (Basel). 2024. PMID: 38674327 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

