SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology
- PMID: 32926098
- PMCID: PMC7488868
- DOI: 10.1084/jem.20201129
SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology
Abstract
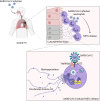
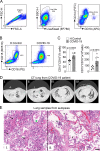
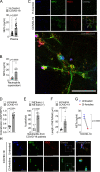
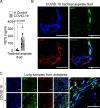
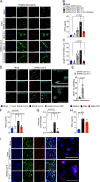
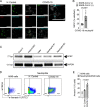
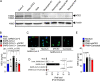
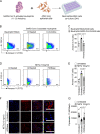
Severe COVID-19 patients develop acute respiratory distress syndrome that may progress to cytokine storm syndrome, organ dysfunction, and death. Considering that neutrophil extracellular traps (NETs) have been described as important mediators of tissue damage in inflammatory diseases, we investigated whether NETs would be involved in COVID-19 pathophysiology. A cohort of 32 hospitalized patients with a confirmed diagnosis of COVID-19 and healthy controls were enrolled. The concentration of NETs was augmented in plasma, tracheal aspirate, and lung autopsies tissues from COVID-19 patients, and their neutrophils released higher levels of NETs. Notably, we found that viable SARS-CoV-2 can directly induce the release of NETs by healthy neutrophils. Mechanistically, NETs triggered by SARS-CoV-2 depend on angiotensin-converting enzyme 2, serine protease, virus replication, and PAD-4. Finally, NETs released by SARS-CoV-2-activated neutrophils promote lung epithelial cell death in vitro. These results unravel a possible detrimental role of NETs in the pathophysiology of COVID-19. Therefore, the inhibition of NETs represents a potential therapeutic target for COVID-19.
© 2020 Veras et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures
References
-
- Altrichter J., Zedler S., Kraft R., Faist E., Mitzner S.R., Sauer M., Windolf J., Scholz M., and Lögters T.. 2010. Neutrophil-derived circulating free DNA (cf-DNA/NETs), a potential prognostic marker for mortality in patients with severe burn injury. Eur. J. Trauma Emerg. Surg. 36:551–557. 10.1007/s00068-010-0013-1 - DOI - PubMed
-
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., et al. ; ACTT-1 Study Group Members . 2020. Remdesivir for the Treatment of Covid-19 - Preliminary Report. N. Engl. J. Med. NEJMoa2007764 10.1056/NEJMoa2007764 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

