Managing a Large-Scale Multiomics Project: A Team Science Case Study in Proteogenomics
- PMID: 32926368
- PMCID: PMC7771375
- DOI: 10.1007/978-1-0716-0849-4_11
Managing a Large-Scale Multiomics Project: A Team Science Case Study in Proteogenomics
Abstract
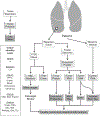
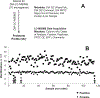
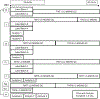
Highly collaborative scientists are often called on to extend their expertise to different types of projects and to expand the scope and scale of projects well beyond their previous experience. For a large-scale project involving "big data" to be successful, several different aspects of the research plan need to be developed and tested, which include but are not limited to the experimental design, sample collection, sample preparation, metadata recording, technical capability, data acquisition, approaches for data analysis, methods for integration of different data types, recruitment of additional expertise as needed to guide the project, and strategies for clear communication throughout the project. To capture this process, we describe an example project in proteogenomics that built on our collective expertise and experience. Key steps included definition of hypotheses, identification of an appropriate clinical cohort, pilot projects to assess feasibility, refinement of experimental designs, and extensive discussions involving the research team throughout the process. The goal of this chapter is to provide the reader with a set of guidelines to support development of other large-scale multiomics projects.
Keywords: Big data; Biostatistics; Cancer; Experimental design; Informatics; Landscape paper; Planning; Proteogenomics.
Figures



Similar articles
-
Proteogenomics: Key Driver for Clinical Discovery and Personalized Medicine.Adv Exp Med Biol. 2016;926:21-47. doi: 10.1007/978-3-319-42316-6_3. Adv Exp Med Biol. 2016. PMID: 27686804 Review.
-
Primary Care Research Team Assessment (PCRTA): development and evaluation.Occas Pap R Coll Gen Pract. 2002 Feb;(81):iii-vi, 1-72. Occas Pap R Coll Gen Pract. 2002. PMID: 12049028 Free PMC article.
-
Bridging the Chromosome-centric and Biology/Disease-driven Human Proteome Projects: Accessible and Automated Tools for Interpreting the Biological and Pathological Impact of Protein Sequence Variants Detected via Proteogenomics.J Proteome Res. 2018 Dec 7;17(12):4329-4336. doi: 10.1021/acs.jproteome.8b00404. Epub 2018 Sep 5. J Proteome Res. 2018. PMID: 30130115
-
Moonshot Objectives: Catalyze New Scientific Breakthroughs-Proteogenomics.Cancer J. 2018 May/Jun;24(3):121-125. doi: 10.1097/PPO.0000000000000315. Cancer J. 2018. PMID: 29794536 Free PMC article. Review.
-
JUMPg: An Integrative Proteogenomics Pipeline Identifying Unannotated Proteins in Human Brain and Cancer Cells.J Proteome Res. 2016 Jul 1;15(7):2309-20. doi: 10.1021/acs.jproteome.6b00344. Epub 2016 Jun 13. J Proteome Res. 2016. PMID: 27225868 Free PMC article.
Cited by
-
Ovarian Cancer: Multi-Omics Data Integration.Int J Mol Sci. 2025 Jun 21;26(13):5961. doi: 10.3390/ijms26135961. Int J Mol Sci. 2025. PMID: 40649740 Free PMC article. Review.
References
-
- Stewart PA, Welsh EA, Slebos RJC, Fang B, Izumi V, Chambers M, Zhang G, Cen L, Pettersson F, Zhang Y, Chen Z, Cheng CH, Thapa R, Thompson Z, Fellows KM, Francis JM, Saller JJ, Mesa T, Zhang C, Yoder S, DeNicola GM, Beg AA, Boyle TA, Teer JK, Ann Chen Y, Koomen JM, Eschrich SA, Haura EB (2019) Proteogenomic landscape of squamous cell lung cancer. Nat Commun 10(1):3578 10.1038/s41467-019-11452-x - DOI - PMC - PubMed
-
- Wilkerson MD, Yin X, Hoadley KA, Liu Y, Hayward MC, Cabanski CR, Muldrew K, Miller CR, Randell SH, Socinski MA, Parsons AM, Funkhouser WK, Lee CB, Roberts PJ, Thorne L, Bernard PS, Perou CM, Hayes DN (2010) Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin Cancer Res 16(19):4864–4875. 10.1158/1078-0432.CCR-10-0199 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

