UHRF1 Is a Novel Druggable Epigenetic Target in Malignant Pleural Mesothelioma
- PMID: 32927122
- PMCID: PMC7775915
- DOI: 10.1016/j.jtho.2020.08.024
UHRF1 Is a Novel Druggable Epigenetic Target in Malignant Pleural Mesothelioma
Abstract
Introduction: Ubiquitin-like with plant homeodomain and ring finger domains 1 (UHRF1) encodes a master regulator of DNA methylation that has emerged as an epigenetic driver in human cancers. To date, no studies have evaluated UHRF1 expression in malignant pleural mesothelioma (MPM). This study was undertaken to explore the therapeutic potential of targeting UHRF1 in MPM.
Methods: Microarray, real-time quantitative reverse transcription-polymerase chain reaction, immunoblot, and immunohistochemistry techniques were used to evaluate UHRF1 expression in normal mesothelial cells (NMCs) cultured with or without asbestos, MPM lines, normal pleura, and primary MPM specimens. The impact of UHRF1 expression on MPM patient survival was evaluated using two independent databases. RNA-sequencing, proliferation, invasion, and colony formation assays, and murine xenograft experiments were performed to evaluate gene expression and growth of MPM cells after biochemical or pharmacologic inhibition of UHRF1 expression.
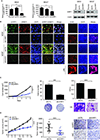
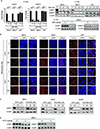
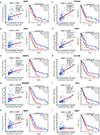
Results: UHRF1 expression was significantly higher in MPM lines and specimens relative to NMC and normal pleura. Asbestos induced UHRF1 expression in NMC. The overexpression of UHRF1 was associated with decreased overall survival in patients with MPM. UHRF1 knockdown reversed genomewide DNA hypomethylation, and inhibited proliferation, invasion, and clonogenicity of MPM cells, and growth of MPM xenografts. These effects were phenocopied by the repurposed chemotherapeutic agent, mithramycin. Biochemical or pharmacologic up-regulation of p53 significantly reduced UHRF1 expression in MPM cells. RNA-sequencing experiments exhibited the pleiotropic effects of UHRF1 down-regulation and identified novel, clinically relevant biomarkers of UHRF1 expression in MPM.
Conclusions: UHRF1 is an epigenetic driver in MPM. These findings support the efforts to target UHRF1 expression or activity for mesothelioma therapy.
Keywords: DNMT1; Epigenetics; Malignant pleural mesothelioma; Prognosis; Therapeutic target; UHRF1.
Copyright © 2020 International Association for the Study of Lung Cancer. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of Interest: None
Figures


Comment in
-
Epigenetic Modifier UHRF1 May Be a Potential Target in Malignant Pleural Mesothelioma.J Thorac Oncol. 2021 Jan;16(1):14-16. doi: 10.1016/j.jtho.2020.10.015. J Thorac Oncol. 2021. PMID: 33384056 No abstract available.
References
-
- Betti M, Aspesi A, Ferrante D, Sculco M, Righi L, Mirabelli D, et al. Sensitivity to asbestos is increased in patients with mesothelioma and pathogenic germline variants in BAP1 or other DNA repair genes. Genes, chromosomes & cancer. 2018;57(11):573–83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

