Identification of Small Molecules that Modulate Mutant p53 Condensation
- PMID: 32927263
- PMCID: PMC7495113
- DOI: 10.1016/j.isci.2020.101517
Identification of Small Molecules that Modulate Mutant p53 Condensation
Abstract
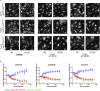
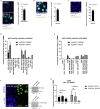
Structural mutants of p53 induce global p53 protein destabilization and misfolding, followed by p53 protein aggregation. First evidence indicates that p53 can be part of protein condensates and that p53 aggregation potentially transitions through a condensate-like state. We show condensate-like states of fluorescently labeled structural mutant p53 in the nucleus of living cancer cells. We furthermore identified small molecule compounds that interact with the p53 protein and lead to dissolution of p53 structural mutant condensates. The same compounds lead to condensation of a fluorescently tagged p53 DNA-binding mutant, indicating that the identified compounds differentially alter p53 condensation behavior depending on the type of p53 mutation. In contrast to p53 aggregation inhibitors, these compounds are active on p53 condensates and do not lead to mutant p53 reactivation. Taken together our study provides evidence for structural mutant p53 condensation in living cells and tools to modulate this process.
Keywords: Biochemistry Methods; Medical Biochemistry; Structural Biology.
© 2020 The Author(s).
Conflict of interest statement
All authors are or were employees of Bayer AG.
Figures





References
-
- Ang H.C., Joerger A.C., Mayer S., Fersht A.R. Effects of common cancer mutations on stability and DNA binding of full-length p53 compared with isolated core domains. J. Biol. Chem. 2006;281:21934–21941. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

