Family Level Phylogenies Reveal Relationships of Plant Viruses within the Order Bunyavirales
- PMID: 32927652
- PMCID: PMC7551631
- DOI: 10.3390/v12091010
Family Level Phylogenies Reveal Relationships of Plant Viruses within the Order Bunyavirales
Abstract
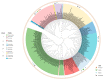
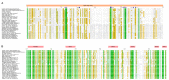
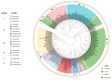
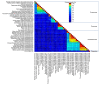
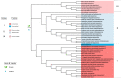
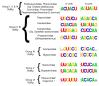
Bunyavirales are negative-sense segmented RNA viruses infecting arthropods, protozoans, plants, and animals. This study examines the phylogenetic relationships of plant viruses within this order, many of which are recently classified species. Comprehensive phylogenetic analyses of the viral RNA dependent RNA polymerase (RdRp), precursor glycoprotein (preGP), the nucleocapsid (N) proteins point toward common progenitor viruses. The RdRp of Fimoviridae and Tospoviridae show a close evolutional relationship while the preGP of Fimoviridae and Phenuiviridae show a closed relationship. The N proteins of Fimoviridae were closer to the Phasmaviridae, the Tospoviridae were close to some Phenuiviridae members and the Peribunyaviridae. The plant viral movement proteins of species within the Tospoviridae and Phenuiviridae were more closely related to each other than to members of the Fimoviridae. Interestingly, distal ends of 3' and 5' untranslated regions of species within the Fimoviridae shared similarity to arthropod and vertebrate infecting members of the Cruliviridae and Peribunyaviridae compared to other plant virus families. Co-phylogeny analysis of the plant infecting viruses indicates that duplication and host switching were more common than co-divergence with a host species.
Keywords: Bunyavirale; RNA virus; cophylogeny; emerging virus; hallmark genes; plant virus; virus evolution.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

