FUT9-Driven Programming of Colon Cancer Cells towards a Stem Cell-Like State
- PMID: 32927726
- PMCID: PMC7565653
- DOI: 10.3390/cancers12092580
FUT9-Driven Programming of Colon Cancer Cells towards a Stem Cell-Like State
Abstract
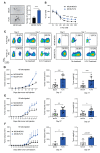
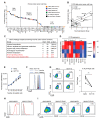
Cancer stem cells (CSCs) are located in dedicated niches, where they remain inert to chemotherapeutic drugs and drive metastasis. Although plasticity in the CSC pool is well appreciated, the molecular mechanisms implicated in the regulation of cancer stemness are still elusive. Here, we define a fucosylation-dependent reprogramming of colon cancer cells towards a stem cell-like phenotype and function. De novo transcriptional activation of Fut9 in the murine colon adenocarcinoma cell line, MC38, followed by RNA seq-based regulon analysis, revealed major gene regulatory networks related to stemness. Lewisx, Sox2, ALDH and CD44 expression, tumorsphere formation, resistance to 5-FU treatment and in vivo tumor growth were increased in FUT9-expressing MC38 cells compared to the control cells. Likewise, human CRC cell lines highly expressing FUT9 displayed phenotypic features of CSCs, which were significantly impaired upon FUT9 knock-out. Finally, in primary CRC FUT9+ tumor cells pathways related to cancer stemness were enriched, providing a clinically meaningful annotation of the complicity of FUT9 in stemness regulation and may open new avenues for therapeutic intervention.
Keywords: colon cancer; drug resistance; fucosylation; glycosylation; pluripotency; stem cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Tsikitis V.L., Malireddy K., Green E.A., Christensen B., Whelan R., Hyder J., Marcello P., Larach S., Lauter D., Sargent D.J., et al. Postoperative surveillance recommendations for early stage colon cancer based on results from the clinical outcomes of surgical therapy trial. J. Clin. Oncol. 2009;27:3671–3676. doi: 10.1200/JCO.2008.20.7050. - DOI - PMC - PubMed
-
- Douillard J.Y., Cunningham D., Roth A.D., Navarro M., James R.D., Karasek P., Jandik P., Iveson T., Carmichael J., Alakl M., et al. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: A multicentre randomised trial. Lancet. 2000;355:1041–1047. doi: 10.1016/S0140-6736(00)02034-1. - DOI - PubMed
-
- Giacchetti S., Perpoint B., Zidani R., Le Bail N., Faggiuolo R., Focan C., Chollet P., Llory J.F., Letourneau Y., Coudert B., et al. Phase III multicenter randomized trial of oxaliplatin added to chronomodulated fluorouracil-leucovorin as first-line treatment of metastatic colorectal cancer. J. Clin. Oncol. 2000;18:136–147. doi: 10.1200/JCO.2000.18.1.136. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

