MasterPATH: network analysis of functional genomics screening data
- PMID: 32928103
- PMCID: PMC7491077
- DOI: 10.1186/s12864-020-07047-2
MasterPATH: network analysis of functional genomics screening data
Abstract
Background: Functional genomics employs several experimental approaches to investigate gene functions. High-throughput techniques, such as loss-of-function screening and transcriptome profiling, allow to identify lists of genes potentially involved in biological processes of interest (so called hit list). Several computational methods exist to analyze and interpret such lists, the most widespread of which aim either at investigating of significantly enriched biological processes, or at extracting significantly represented subnetworks.
Results: Here we propose a novel network analysis method and corresponding computational software that employs the shortest path approach and centrality measure to discover members of molecular pathways leading to the studied phenotype, based on functional genomics screening data. The method works on integrated interactomes that consist of both directed and undirected networks - HIPPIE, SIGNOR, SignaLink, TFactS, KEGG, TransmiR, miRTarBase. The method finds nodes and short simple paths with significant high centrality in subnetworks induced by the hit genes and by so-called final implementers - the genes that are involved in molecular events responsible for final phenotypic realization of the biological processes of interest. We present the application of the method to the data from miRNA loss-of-function screen and transcriptome profiling of terminal human muscle differentiation process and to the gene loss-of-function screen exploring the genes that regulates human oxidative DNA damage recognition. The analysis highlighted the possible role of several known myogenesis regulatory miRNAs (miR-1, miR-125b, miR-216a) and their targets (AR, NR3C1, ARRB1, ITSN1, VAV3, TDGF1), as well as linked two major regulatory molecules of skeletal myogenesis, MYOD and SMAD3, to their previously known muscle-related targets (TGFB1, CDC42, CTCF) and also to a number of proteins such as C-KIT that have not been previously studied in the context of muscle differentiation. The analysis also showed the role of the interaction between H3 and SETDB1 proteins for oxidative DNA damage recognition.
Conclusion: The current work provides a systematic methodology to discover members of molecular pathways in integrated networks using functional genomics screening data. It also offers a valuable instrument to explain the appearance of a set of genes, previously not associated with the process of interest, in the hit list of each particular functional genomics screening.
Keywords: Centrality; DNA repair; Loss-of-function screening; Molecular pathway; Muscle differentiation; Network analysis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
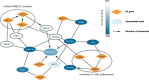

Similar articles
-
A Map of the microRNA Regulatory Networks Identified by Experimentally Validated microRNA-Target Interactions in Five Domestic Animals: Cattle, Pig, Sheep, Dog, and Chicken.OMICS. 2019 Sep;23(9):448-456. doi: 10.1089/omi.2019.0082. Epub 2019 Aug 5. OMICS. 2019. PMID: 31381467
-
An integrated analysis of mRNA and miRNA in skeletal muscle from myostatin-edited Meishan pigs.Genome. 2019 May;62(5):305-315. doi: 10.1139/gen-2018-0110. Epub 2019 Mar 26. Genome. 2019. PMID: 30913397
-
Integrated analyses to reconstruct microRNA-mediated regulatory networks in mouse liver using high-throughput profiling.BMC Genomics. 2015;16 Suppl 2(Suppl 2):S12. doi: 10.1186/1471-2164-16-S2-S12. Epub 2015 Jan 21. BMC Genomics. 2015. PMID: 25707768 Free PMC article.
-
miRNA-miRNA crosstalk: from genomics to phenomics.Brief Bioinform. 2017 Nov 1;18(6):1002-1011. doi: 10.1093/bib/bbw073. Brief Bioinform. 2017. PMID: 27551063 Review.
-
Reverse-engineering human regulatory networks.Wiley Interdiscip Rev Syst Biol Med. 2012 Jul-Aug;4(4):311-25. doi: 10.1002/wsbm.1159. Epub 2012 Jan 13. Wiley Interdiscip Rev Syst Biol Med. 2012. PMID: 22246697 Free PMC article. Review.
Cited by
-
A Resource for the Network Representation of Cell Perturbations Caused by SARS-CoV-2 Infection.Genes (Basel). 2021 Mar 22;12(3):450. doi: 10.3390/genes12030450. Genes (Basel). 2021. PMID: 33809949 Free PMC article.
-
Mechanism and Utilization of Ogura Cytoplasmic Male Sterility in Cruciferae Crops.Int J Mol Sci. 2022 Aug 13;23(16):9099. doi: 10.3390/ijms23169099. Int J Mol Sci. 2022. PMID: 36012365 Free PMC article. Review.
-
Emerging landscape of molecular interaction networks:Opportunities, challenges and prospects.J Biosci. 2022;47(2):24. doi: 10.1007/s12038-022-00253-y. J Biosci. 2022. PMID: 36210749 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

