CrustyBase: an interactive online database for crustacean transcriptomes
- PMID: 32928113
- PMCID: PMC7490944
- DOI: 10.1186/s12864-020-07063-2
CrustyBase: an interactive online database for crustacean transcriptomes
Abstract
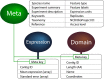
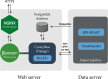
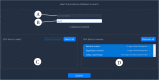
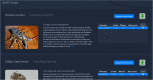
Transcriptome sequencing has opened the field of genomics to a wide variety of researchers, owing to its efficiency, applicability across species and ability to quantify gene expression. The resulting datasets are a rich source of information that can be mined for many years into the future, with each dataset providing a unique angle on a specific context in biology. Maintaining accessibility to this accumulation of data presents quite a challenge for researchers.The primary focus of conventional genomics databases is the storage, navigation and interpretation of sequence data, which is typically classified down to the level of a species or individual. The addition of expression data adds a new dimension to this paradigm - the sampling context. Does gene expression describe different tissues, a temporal distribution or an experimental treatment? These data not only describe an individual, but the biological context surrounding that individual. The structure and utility of a transcriptome database must therefore reflect these attributes. We present an online database which has been designed to maximise the accessibility of crustacean transcriptome data by providing intuitive navigation within and between datasets and instant visualization of gene expression and protein structure.The site is accessible at https://crustybase.org and currently holds 10 datasets from a range of crustacean species. It also allows for upload of novel transcriptome datasets through a simple web interface, allowing the research community to contribute their own data to a pool of shared knowledge.
Keywords: Crab; Crayfish; Expression; Gene; Genomics; Lobster; RNA-seq; Search; Shrimp; Visualize.
Conflict of interest statement
None to declare.
Figures

References
-
- National Centre for Biotechnology Information National Library of Medicinehttps://ncbi.nlm.nih.gov Accessed 01 Nov 2019.
-
- Nguyen TV, Jung H, Rotllant G, Hurwood D, Mather P, Ventura T. Guidelines for RNA-seq projects: applications and opportunities in non-model decapod crustacean species. Hydrobiologia. 2018;825(1):5–27. doi: 10.1007/s10750-018-3682-0. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

