Genetic diversity and different cross-neutralization capability of porcine circovirus type 2 isolates recently circulating in South Korea
- PMID: 32928247
- PMCID: PMC7488706
- DOI: 10.1186/s12917-020-02549-3
Genetic diversity and different cross-neutralization capability of porcine circovirus type 2 isolates recently circulating in South Korea
Abstract
Background: Porcine circovirus type 2 (PCV2) is a small single-stranded DNA virus and a primary cause of PCV-associated diseases (PCVAD) that result insubstantial economic loss for swine farms. Between 2016 and 2018, PCV2 field viruses were isolated from PCVAD-affected swine farms in South Korea and investigated for genetic and antigenic heterogeneity.
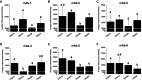
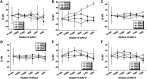
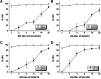
Results: The genetic analysis of ORF2 showed that the genotype of the Korean PCV2 field isolates has been rapidly shifted from PCV2a or 2b to mutant PCV2b known as PCV2d with 82.6 to 100% amino acid sequence similarity. PCV2-specific monoclonal antibodies (mAbs) demonstrated variable antigen-binding activity to four representative Korean PCV2 field isolates [QIA215 (PCV2a), QIA418 (PCV2b), QIA169 (PCV2d), and QIA244 (PCV2d)] without genotype specificity, and one mAb showed neutralization activity to QIA215. In a cross-virus neutralization assay using anti-PCV2 sera of pigs and guinea pigs injected with a commercial vaccine and the Korean PCV2 field isolates, the anti-porcine sera of a commercial vaccine had high neutralization activity against QIA215 and QIA418 with statistically lower activity against PCV2d viruses. Anti-guinea pig sera of QIA215, QIA418, QIA169, and a commercial vaccine had high neutralization activity against all of the viruses with significantly lower activity against QIA244. Importantly, anti-guinea pig sera of QIA244 had high neutralization activity against all of the viruses.
Conclusions: This study confirmed genetic and antigenic diversity among recent PCV2 field isolates in Korean swine farms, and the strain-based difference in virus neutralization capability should be considered for more effective control by vaccination.
Keywords: Antigenic diversity; Genotype; Neutralization; Porcine circovirus type 2.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Evaluation of a porcine circovirus type 2a (PCV2a) vaccine efficacy against experimental PCV2a, PCV2b, and PCV2d challenge.Vet Microbiol. 2019 Apr;231:87-92. doi: 10.1016/j.vetmic.2019.03.002. Epub 2019 Mar 5. Vet Microbiol. 2019. PMID: 30955830
-
A chimeric virus created by DNA shuffling of the capsid genes of different subtypes of porcine circovirus type 2 (PCV2) in the backbone of the non-pathogenic PCV1 induces protective immunity against the predominant PCV2b and the emerging PCV2d in pigs.Virology. 2016 Nov;498:82-93. doi: 10.1016/j.virol.2016.08.011. Epub 2016 Aug 24. Virology. 2016. PMID: 27564544
-
A Porcine circovirus type 2b (PCV2b)-based experimental vaccine is effective in the PCV2b-Mycoplasma hyopneumoniae coinfection pig model.Vaccine. 2019 Oct 16;37(44):6688-6695. doi: 10.1016/j.vaccine.2019.09.029. Epub 2019 Sep 16. Vaccine. 2019. PMID: 31537445
-
Porcine Circovirus Type 2 (PCV2) Vaccines in the Context of Current Molecular Epidemiology.Viruses. 2017 May 6;9(5):99. doi: 10.3390/v9050099. Viruses. 2017. PMID: 28481275 Free PMC article. Review.
-
Genetic variation of porcine circovirus type 2 (PCV2) and its relevance to vaccination, pathogenesis and diagnosis.Virus Res. 2012 Mar;164(1-2):68-77. doi: 10.1016/j.virusres.2011.11.018. Epub 2011 Dec 17. Virus Res. 2012. PMID: 22198217 Review.
Cited by
-
Porcine Circovirus (PCV) Genotype 2d-Based Virus-like Particles (VLPs) Induced Broad Cross-Neutralizing Antibodies against Diverse Genotypes and Provided Protection in Dual-Challenge Infection of a PCV2d Virus and a Type 1 Porcine Reproductive and Respiratory Syndrome Virus (PRRSV).Pathogens. 2021 Sep 6;10(9):1145. doi: 10.3390/pathogens10091145. Pathogens. 2021. PMID: 34578177 Free PMC article.
-
A Field Efficacy Trial of Recombinant Porcine Circovirus Type 2d Vaccine in Three Herds.Vaccines (Basel). 2023 Sep 16;11(9):1497. doi: 10.3390/vaccines11091497. Vaccines (Basel). 2023. PMID: 37766173 Free PMC article.
-
Porcine Circovirus 2 Genotypes, Immunity and Vaccines: Multiple Genotypes but One Single Serotype.Pathogens. 2020 Dec 14;9(12):1049. doi: 10.3390/pathogens9121049. Pathogens. 2020. PMID: 33327478 Free PMC article. Review.
-
An updated phylogeography and population dynamics of porcine circovirus 2 genotypes: are they reaching an equilibrium?Front Microbiol. 2024 Oct 29;15:1500498. doi: 10.3389/fmicb.2024.1500498. eCollection 2024. Front Microbiol. 2024. PMID: 39534503 Free PMC article.
-
Epidemiological and genetic variation analysis of emerging porcine circovirus type 2 in Henan Province, 2023.Front Vet Sci. 2025 Apr 22;12:1598383. doi: 10.3389/fvets.2025.1598383. eCollection 2025. Front Vet Sci. 2025. PMID: 40331216 Free PMC article.
References
-
- Segales J, Olvera A, Grau-Roma L, Charreyre C, Nauwynck H, Larsen L, Dupont K, McCullough K, Ellis J, Krakowka S, Mankertz A, Fredholm M, Fossum C, Timmusk S, Stockhofe-Zurwieden N, Beattie V, Armstrong D, Grassland B, Baekbo P, Allan G. PCV-2 genotype definition and nomenclature. Vet Rec. 2008;162(26):867–868. doi: 10.1136/vr.162.26.867. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

