Emerging HxNy Influenza A Viruses
- PMID: 32928891
- PMCID: PMC8805644
- DOI: 10.1101/cshperspect.a038406
Emerging HxNy Influenza A Viruses
Abstract
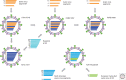
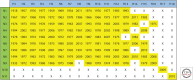
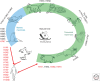
The continuous emergence and reemergence of diverse subtypes of influenza A viruses, which are known as "HxNy" and are mediated through the reassortment of viral genomes, account for seasonal epidemics, occasional pandemics, and zoonotic outbreaks. We summarize and discuss the characteristics of historic human pandemic HxNy viruses and diverse subtypes of HxNy among wild birds, mammals, and live poultry markets. In addition, we summarize the key molecular features of emerging infectious HxNy influenza viruses from the perspectives of the receptor binding of Hx, the inhibitor-binding specificities and drug-resistance features of Ny, and the matching of the gene segments. Our work enhances our understanding of the potential threats of novel reassortant influenza viruses to public health and provides recommendations for effective prevention, control, and research of this pathogen.
Copyright © 2022 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
