Ferric uptake regulator (Fur) reversibly binds a [2Fe-2S] cluster to sense intracellular iron homeostasis in Escherichia coli
- PMID: 32928958
- PMCID: PMC7667960
- DOI: 10.1074/jbc.RA120.014814
Ferric uptake regulator (Fur) reversibly binds a [2Fe-2S] cluster to sense intracellular iron homeostasis in Escherichia coli
Abstract
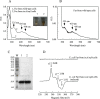
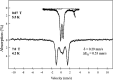
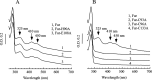
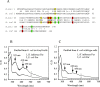
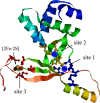
The ferric uptake regulator (Fur) is a global transcription factor that regulates intracellular iron homeostasis in bacteria. The current hypothesis states that when the intracellular "free" iron concentration is elevated, Fur binds ferrous iron, and the iron-bound Fur represses the genes encoding for iron uptake systems and stimulates the genes encoding for iron storage proteins. However, the "iron-bound" Fur has never been isolated from any bacteria. Here we report that the Escherichia coli Fur has a bright red color when expressed in E. coli mutant cells containing an elevated intracellular free iron content because of deletion of the iron-sulfur cluster assembly proteins IscA and SufA. The acid-labile iron and sulfide content analyses in conjunction with the EPR and Mössbauer spectroscopy measurements and the site-directed mutagenesis studies show that the red Fur protein binds a [2Fe-2S] cluster via conserved cysteine residues. The occupancy of the [2Fe-2S] cluster in Fur protein is ∼31% in the E. coli iscA/sufA mutant cells and is decreased to ∼4% in WT E. coli cells. Depletion of the intracellular free iron content using the membrane-permeable iron chelator 2,2´-dipyridyl effectively removes the [2Fe-2S] cluster from Fur in E. coli cells, suggesting that Fur senses the intracellular free iron content via reversible binding of a [2Fe-2S] cluster. The binding of the [2Fe-2S] cluster in Fur appears to be highly conserved, because the Fur homolog from Hemophilus influenzae expressed in E. coli cells also reversibly binds a [2Fe-2S] cluster to sense intracellular iron homeostasis.
Keywords: Escherichia coli (E. coli); IscA; Mossbauer spectroscopy; electron paramagnetic resonance (EPR); ferric uptake regulator (Fur); intracellular iron homeostasis; iron homeostasis; iron metabolism; iron–sulfur cluster; iron–sulfur protein; repressor protein.
© 2020 Fontenot et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures


Comment in
-
Do FeS clusters rule bacterial iron regulation?J Biol Chem. 2020 Nov 13;295(46):15464-15465. doi: 10.1074/jbc.H120.016190. J Biol Chem. 2020. PMID: 33188081 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

