Exploring the landscape of model representations
- PMID: 32929015
- PMCID: PMC7533877
- DOI: 10.1073/pnas.2000098117
Exploring the landscape of model representations
Abstract
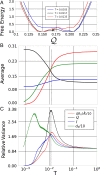
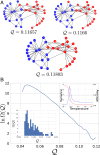
The success of any physical model critically depends upon adopting an appropriate representation for the phenomenon of interest. Unfortunately, it remains generally challenging to identify the essential degrees of freedom or, equivalently, the proper order parameters for describing complex phenomena. Here we develop a statistical physics framework for exploring and quantitatively characterizing the space of order parameters for representing physical systems. Specifically, we examine the space of low-resolution representations that correspond to particle-based coarse-grained (CG) models for a simple microscopic model of protein fluctuations. We employ Monte Carlo (MC) methods to sample this space and determine the density of states for CG representations as a function of their ability to preserve the configurational information, I, and large-scale fluctuations, Q, of the microscopic model. These two metrics are uncorrelated in high-resolution representations but become anticorrelated at lower resolutions. Moreover, our MC simulations suggest an emergent length scale for coarse-graining proteins, as well as a qualitative distinction between good and bad representations of proteins. Finally, we relate our work to recent approaches for clustering graphs and detecting communities in networks.
Keywords: entropy; information theory; multiscale modeling; networks; proteins.
Conflict of interest statement
The authors declare no competing interest.
Figures




References
-
- Goldenfeld N., Kadanoff L. P., Simple lessons from complexity. Science 284, 87–89 (1999). - PubMed
-
- Callen H. B., Thermodynamics and an Introduction to Thermostatistics (Wiley, 1985).
-
- Levitt M., Warshel A., Computer simulation of protein folding. Nature 253, 694–698 (1975). - PubMed
-
- Peter C., Kremer K., Multiscale simulation of soft matter systems. Faraday Discuss. 144, 9–24 (2010). - PubMed
-
- Noid W. G., Perspective: Coarse-grained models for biomolecular systems. J. Chem. Phys. 139, 090901 (2013). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

