Small-Molecule Inhibitor of 8-Oxoguanine DNA Glycosylase 1 Regulates Inflammatory Responses during Pseudomonas aeruginosa Infection
- PMID: 32929043
- PMCID: PMC7541742
- DOI: 10.4049/jimmunol.1901533
Small-Molecule Inhibitor of 8-Oxoguanine DNA Glycosylase 1 Regulates Inflammatory Responses during Pseudomonas aeruginosa Infection
Abstract
The DNA repair enzyme 8-oxoguanine DNA glycosylase 1 (OGG1), which excises 8-oxo-7,8-dihydroguanine lesions induced in DNA by reactive oxygen species, has been linked to the pathogenesis of lung diseases associated with bacterial infections. A recently developed small molecule, SU0268, has demonstrated selective inhibition of OGG1 activity; however, its role in attenuating inflammatory responses has not been tested. In this study, we report that SU0268 has a favorable effect on bacterial infection both in mouse alveolar macrophages (MH-S cells) and in C57BL/6 wild-type mice by suppressing inflammatory responses, particularly promoting type I IFN responses. SU0268 inhibited proinflammatory responses during Pseudomonas aeruginosa (PA14) infection, which is mediated by the KRAS-ERK1-NF-κB signaling pathway. Furthermore, SU0268 induces the release of type I IFN by the mitochondrial DNA-cGAS-STING-IRF3-IFN-β axis, which decreases bacterial loads and halts disease progression. Collectively, our results demonstrate that the small-molecule inhibitor of OGG1 (SU0268) can attenuate excessive inflammation and improve mouse survival rates during PA14 infection. This strong anti-inflammatory feature may render the inhibitor as an alternative treatment for controlling severe inflammatory responses to bacterial infection.
Copyright © 2020 by The American Association of Immunologists, Inc.
Conflict of interest statement
Footnotes
Disclosures
The authors have no financial conflicts of interest.
Figures
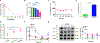
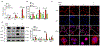





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

