The ERα-miR-575-p27 feedback loop regulates tamoxifen sensitivity in ER-positive Breast Cancer
- PMID: 32929377
- PMCID: PMC7482812
- DOI: 10.7150/thno.46297
The ERα-miR-575-p27 feedback loop regulates tamoxifen sensitivity in ER-positive Breast Cancer
Abstract
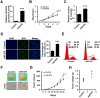
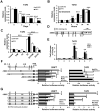
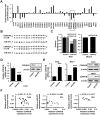
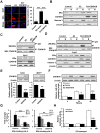
Background: Breast cancer is the most common malignancy, and approximately 70% of breast cancers are estrogen receptor-α (ERα) positive. The anti-estrogen tamoxifen is a highly effective and commonly used treatment for patients with ER+ breast cancer. However, 30% of breast cancer patients fail adjuvant tamoxifen therapy and most of metastatic breast cancer patients develop tamoxifen resistance. Although increasing evidence suggests that microRNA (miRNA) dysregulation influences tamoxifen sensitivity, the mechanism of the cross-talk between miRNA and ERα signaling remains unclear. miR-575 has been reported to be involved in carcinogenesis and progression, however, the role of miR-575 in breast cancer remains limited. The aim of this study was to understand the mechanism of miR-575 in breast cancer tamoxifen resistance. Method: RT-qPCR was employed to assess miR-575 expression in breast cancer tissues and cell lines. The association of miR-575 expression with overall survival in patients with breast cancer was evaluated with KM plotter. Additionally, the effects of miR-575 on breast cancer proliferation and tamoxifen sensitivity were investigated both in vitro and in vivo. Bioinformatic analyses and luciferase reporter assays were performed to validate CDKN1B and BRCA1 as direct targets of miR-31-5p. The ERα binding sites in the miR-575 promoter region was validated with ChIP and luciferase assays. ERα interactions with CDKN1B, cyclin D1 or BRCA1 were determined by IP analysis, and protein expression levels and localization were analyzed by western blotting and immunofluorescence, respectively. Results: miR-575 levels were higher in ER+ breast cancer than in ER- breast cancer and patients with high miR-575 expression had a significantly poorer outcome than those with low miR-575 expression. ERα bound the miR-575 promoter to activate its transcription, and tamoxifen treatment downregulated miR-575 expression in ER+ breast cancer. Overexpression of miR-575 decreased tamoxifen sensitivity by targeting CDKN1B and BRCA1. CDKN1B and BRCA1 were both able to antagonize ERα activity by inhibiting ERα nuclear translocation and interaction with cyclin D1. Furthermore, miR-575 expression was found to be upregulated in ER+ breast cancer cell with acquired tamoxifen resistance, whereas depletion of miR-575 partially re-sensitized these cells to tamoxifen by regulation of CDKN1B. Conclusions: Our data reveal the ERα-miR-575-CDKN1B feedback loop in ER+ breast cancer, suggesting that miR-575 can be used as a prognostic biomarker in patients with ER+ breast cancer, as well as a predictor or a promising target for tamoxifen sensitivity.
Keywords: miR-575 regulates ER+ breast cancer tamoxifen sensitivity.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



Similar articles
-
Decreased BRCA1 confers tamoxifen resistance in breast cancer cells by altering estrogen receptor-coregulator interactions.Oncogene. 2009 Jan 29;28(4):575-86. doi: 10.1038/onc.2008.405. Epub 2008 Nov 10. Oncogene. 2009. PMID: 18997820 Free PMC article.
-
MiRNA-27a sensitizes breast cancer cells to treatment with Selective Estrogen Receptor Modulators.Breast. 2019 Feb;43:31-38. doi: 10.1016/j.breast.2018.10.007. Epub 2018 Oct 18. Breast. 2019. PMID: 30415143
-
Loss of Estrogen-Regulated MIR135A1 at 3p21.1 Promotes Tamoxifen Resistance in Breast Cancer.Cancer Res. 2018 Sep 1;78(17):4915-4928. doi: 10.1158/0008-5472.CAN-18-0069. Epub 2018 Jun 26. Cancer Res. 2018. PMID: 29945962
-
Identification of miRNAs as biomarkers for acquired endocrine resistance in breast cancer.Mol Cell Endocrinol. 2017 Nov 15;456:76-86. doi: 10.1016/j.mce.2017.02.004. Epub 2017 Feb 3. Mol Cell Endocrinol. 2017. PMID: 28163101 Review.
-
Crosstalk of methylation and tamoxifen in breast cancer (Review).Mol Med Rep. 2024 Oct;30(4):180. doi: 10.3892/mmr.2024.13304. Epub 2024 Aug 12. Mol Med Rep. 2024. PMID: 39129315 Free PMC article. Review.
Cited by
-
ZNF384-ZEB1 feedback loop regulates breast cancer metastasis.Mol Med. 2022 Sep 13;28(1):111. doi: 10.1186/s10020-022-00541-1. Mol Med. 2022. PMID: 36100877 Free PMC article.
-
YY1 transcription factor induces proliferation and aerobic glycolysis of neuroblastoma cells via LDHA regulation.Exp Ther Med. 2022 Nov 29;25(1):37. doi: 10.3892/etm.2022.11736. eCollection 2023 Jan. Exp Ther Med. 2022. PMID: 36569438 Free PMC article.
-
MIR497HG-Derived miR-195 and miR-497 Mediate Tamoxifen Resistance via PI3K/AKT Signaling in Breast Cancer.Adv Sci (Weinh). 2023 Apr;10(12):e2204819. doi: 10.1002/advs.202204819. Epub 2023 Feb 23. Adv Sci (Weinh). 2023. PMID: 36815359 Free PMC article.
-
LINC01189-miR-586-ZEB1 feedback loop regulates breast cancer progression through Wnt/β-catenin signaling pathway.Mol Ther Nucleic Acids. 2021 Jun 24;25:455-467. doi: 10.1016/j.omtn.2021.06.007. eCollection 2021 Sep 3. Mol Ther Nucleic Acids. 2021. PMID: 34513288 Free PMC article.
-
Obesity as a Risk Factor for Breast Cancer-The Role of miRNA.Int J Mol Sci. 2022 Dec 10;23(24):15683. doi: 10.3390/ijms232415683. Int J Mol Sci. 2022. PMID: 36555323 Free PMC article. Review.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

