LncRNA DSCAM-AS1 interacts with YBX1 to promote cancer progression by forming a positive feedback loop that activates FOXA1 transcription network
- PMID: 32929382
- PMCID: PMC7482804
- DOI: 10.7150/thno.47830
LncRNA DSCAM-AS1 interacts with YBX1 to promote cancer progression by forming a positive feedback loop that activates FOXA1 transcription network
Abstract
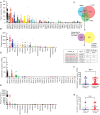
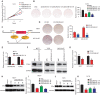
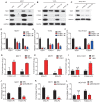
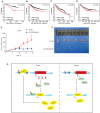
Rationale: The forkhead box A1 (FOXA1) is a crucial transcription factor in initiation and development of breast, lung and prostate cancer. Previous studies about the FOXA1 transcriptional network were mainly focused on protein-coding genes. Its regulatory network of long non-coding RNAs (lncRNAs) and their role in FOXA1 oncogenic activity remains unknown. Methods: The Cancer Genome Atlas (TCGA) data, RNA-seq and ChIP-seq data were used to analyze FOXA1 regulated lncRNAs. RT-qPCR was used to detect the expression of DSCAM-AS1, RT-qPCR and Western blotting were used to determine the expression of FOXA1, estrogen receptor α (ERα) and Y box binding protein 1 (YBX1). RNA pull-down and RIP-qPCR were employed to investigate the interaction between DSCAM-AS1 and YBX1. The effect of DSCAM-AS1 on malignant phenotypes was examined through in vitro and in vivo assays. Results: In this study, we conducted a global analysis of FOXA1 regulated lncRNAs. For detailed analysis, we chose lncRNA DSCAM-AS1, which is specifically expressed in lung adenocarcinoma, breast and prostate cancer. The expression level of DSCAM-AS1 is regulated by two super-enhancers (SEs) driven by FOXA1. High expression levels of DSCAM-AS1 was associated with poor prognosis. Knockout experiments showed DSCAM-AS1 was essential for the growth of xenograft tumors. Moreover, we demonstrated DSCAM-AS1 can regulate the expression of the master transcriptional factor FOXA1. In breast cancer, DSCAM-AS1 was also found to regulate ERα. Mechanistically, DSCAM-AS1 interacts with YBX1 and influences the recruitment of YBX1 in the promoter regions of FOXA1 and ERα. Conclusion: Our study demonstrated that lncRNA DSCAM-AS1 was transcriptionally activated by super-enhancers driven by FOXA1 and exhibited lineage-specific expression pattern. DSCAM-AS1 can promote cancer progression by interacting with YBX1 and regulating expression of FOXA1 and ERα.
Keywords: ERα; FOXA1; breast cancer; lncRNAs; lung adenocarcinoma; super-enhancer.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures


Similar articles
-
A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis.Mol Cancer. 2019 Feb 19;18(1):28. doi: 10.1186/s12943-019-0957-7. Mol Cancer. 2019. PMID: 30782188 Free PMC article.
-
YBX1/lncRNA SBF2-AS1 interaction regulates proliferation and tamoxifen sensitivity via PI3K/AKT/MTOR signaling in breast cancer cells.Mol Biol Rep. 2023 Apr;50(4):3413-3428. doi: 10.1007/s11033-023-08308-5. Epub 2023 Feb 8. Mol Biol Rep. 2023. PMID: 36754932
-
Luminal lncRNAs Regulation by ERα-Controlled Enhancers in a Ligand-Independent Manner in Breast Cancer Cells.Int J Mol Sci. 2018 Feb 16;19(2):593. doi: 10.3390/ijms19020593. Int J Mol Sci. 2018. PMID: 29462945 Free PMC article.
-
LncRNA DSCAM-AS1: A Pivotal Therapeutic Target in Cancer.Mini Rev Med Chem. 2023;23(5):530-536. doi: 10.2174/1389557522666220822121935. Mini Rev Med Chem. 2023. PMID: 35996247 Review.
-
FOXA1: a transcription factor with parallel functions in development and cancer.Biosci Rep. 2012 Apr 1;32(2):113-30. doi: 10.1042/BSR20110046. Biosci Rep. 2012. PMID: 22115363 Free PMC article. Review.
Cited by
-
EpCAM as a Novel Biomarker for Survivals in Prostate Cancer Patients.Front Cell Dev Biol. 2022 Apr 20;10:843604. doi: 10.3389/fcell.2022.843604. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35517503 Free PMC article.
-
FOXA1-induced LINC00621 promotes lung adenocarcinoma progression via activating the TGF-β signaling pathway.Thorac Cancer. 2023 Jul;14(21):2026-2037. doi: 10.1111/1759-7714.14986. Epub 2023 Jun 5. Thorac Cancer. 2023. PMID: 37277890 Free PMC article.
-
S1PR1 suppresses lung adenocarcinoma progression through p-STAT1/miR-30c-5 p/FOXA1 pathway.J Exp Clin Cancer Res. 2024 Nov 18;43(1):304. doi: 10.1186/s13046-024-03230-5. J Exp Clin Cancer Res. 2024. PMID: 39551792 Free PMC article.
-
M6A-mediated upregulation of lncRNA TUG1 in liver cancer cells regulates the antitumor response of CD8+ T cells and phagocytosis of macrophages.Adv Sci (Weinh). 2024 Sep;11(34):e2400695. doi: 10.1002/advs.202400695. Epub 2024 Jul 9. Adv Sci (Weinh). 2024. PMID: 38981064 Free PMC article.
-
Knockdown of lncRNA ACTA2-AS1 reverses cisplatin resistance of ovarian cancer cells via inhibition of miR-378a-3p-regulated Wnt5a.Bioengineered. 2022 Apr;13(4):9829-9838. doi: 10.1080/21655979.2022.2061181. Bioengineered. 2022. PMID: 35412951 Free PMC article.
References
-
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–89. - PubMed
-
- Gao N, Ishii K, Mirosevich J, Kuwajima S, Oppenheimer SR, Roberts RL. et al. Forkhead box A1 regulates prostate ductal morphogenesis and promotes epithelial cell maturation. Development. 2005;132:3431–43. - PubMed
-
- Wan H, Dingle S, Xu Y, Besnard V, Kaestner KH, Ang SL. et al. Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J Biol Chem. 2005;280:13809–16. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

