locStra: Fast analysis of regional/global stratification in whole-genome sequencing studies
- PMID: 32929743
- PMCID: PMC7856019
- DOI: 10.1002/gepi.22356
locStra: Fast analysis of regional/global stratification in whole-genome sequencing studies
Abstract
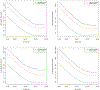
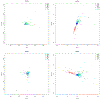
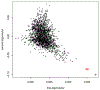
locStra is an -package for the analysis of regional and global population stratification in whole-genome sequencing (WGS) studies, where regional stratification refers to the substructure defined by the loci in a particular region on the genome. Population substructure can be assessed based on the genetic covariance matrix, the genomic relationship matrix, and the unweighted/weighted genetic Jaccard similarity matrix. Using a sliding window approach, the regional similarity matrices are compared with the global ones, based on user-defined window sizes and metrics, for example, the correlation between regional and global eigenvectors. An algorithm for the specification of the window size is provided. As the implementation fully exploits sparse matrix algebra and is written in C++, the analysis is highly efficient. Even on single cores, for realistic study sizes (several thousand subjects, several million rare variants per subject), the runtime for the genome-wide computation of all regional similarity matrices does typically not exceed one hour, enabling an unprecedented investigation of regional stratification across the entire genome. The package is applied to three WGS studies, illustrating the varying patterns of regional substructure across the genome and its beneficial effects on association testing.
Keywords: population stratification; population substructure; regional analysis; similarity matrix; whole-genome sequencing.
© 2020 Wiley Periodicals LLC.
Conflict of interest statement
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
Figures


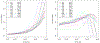



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

