Single-Cell RNA Sequencing Unveils Unique Transcriptomic Signatures of Organ-Specific Endothelial Cells
- PMID: 32929989
- PMCID: PMC7658053
- DOI: 10.1161/CIRCULATIONAHA.119.041433
Single-Cell RNA Sequencing Unveils Unique Transcriptomic Signatures of Organ-Specific Endothelial Cells
Abstract
Background: Endothelial cells (ECs) display considerable functional heterogeneity depending on the vessel and tissue in which they are located. Whereas these functional differences are presumably imprinted in the transcriptome, the pathways and networks that sustain EC heterogeneity have not been fully delineated.
Methods: To investigate the transcriptomic basis of EC specificity, we analyzed single-cell RNA sequencing data from tissue-specific mouse ECs generated by the Tabula Muris consortium. We used a number of bioinformatics tools to uncover markers and sources of EC heterogeneity from single-cell RNA sequencing data.
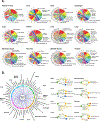
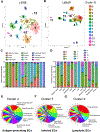
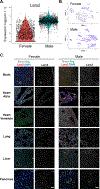
Results: We found a strong correlation between tissue-specific EC transcriptomic measurements generated by either single-cell RNA sequencing or bulk RNA sequencing, thus validating the approach. Using a graph-based clustering algorithm, we found that certain tissue-specific ECs cluster strongly by tissue (eg, liver, brain), whereas others (ie, adipose, heart) have considerable transcriptomic overlap with ECs from other tissues. We identified novel markers of tissue-specific ECs and signaling pathways that may be involved in maintaining their identity. Sex was a considerable source of heterogeneity in the endothelial transcriptome and we discovered Lars2 to be a gene that is highly enriched in ECs from male mice. We found that markers of heart and lung ECs in mice were conserved in human fetal heart and lung ECs. We identified potential angiocrine interactions between tissue-specific ECs and other cell types by analyzing ligand and receptor expression patterns.
Conclusions: We used single-cell RNA sequencing data generated by the Tabula Muris consortium to uncover transcriptional networks that maintain tissue-specific EC identity and to identify novel angiocrine and functional relationships between tissue-specific ECs.
Keywords: computational biology; endothelial cells; sequence analysis, RNA.
Conflict of interest statement
DISCLOSURES
Dr. Wu is a co-founder of Khloris Biosciences but has no competing interests, as the work presented here is completely independent. The other authors declare no competing interests.
Figures




References
-
- Cines DB, Pollak ES, Buck CA, Loscalzo J, Zimmerman GA, McEver RP, Pober JS, Wick TM, Konkle BA, Schwartz BS, Barnathan ES, McCrae KR, Hug BA, Schmidt AM, Stern DM. Endothelial cells in physiology and in the pathophysiology of vascular disorders. Blood. 1998;91:3527–3561. - PubMed
-
- Vanhoutte PM, Shimokawa H, Feletou M, Tang EHC. Endothelial dysfunction and vascular disease - a 30th anniversary update. Acta Physiol Oxf Engl. 2017;219:22–96. - PubMed
-
- Rohlenova K, Goveia J, García-Caballero M, Subramanian A, Kalucka J, Treps L, Falkenberg KD, de Rooij LPMH, Zheng Y, Lin L, et al. Single-cell RNA sequencing maps endothelial metabolic plasticity in pathological angiogenesis. Cell Metab 2020;31:862–877.e14. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

