Resilience of clinical text de-identified with "hiding in plain sight" to hostile reidentification attacks by human readers
- PMID: 32930712
- PMCID: PMC7647331
- DOI: 10.1093/jamia/ocaa095
Resilience of clinical text de-identified with "hiding in plain sight" to hostile reidentification attacks by human readers
Abstract
Objective: Effective, scalable de-identification of personally identifying information (PII) for information-rich clinical text is critical to support secondary use, but no method is 100% effective. The hiding-in-plain-sight (HIPS) approach attempts to solve this "residual PII problem." HIPS replaces PII tagged by a de-identification system with realistic but fictitious (resynthesized) content, making it harder to detect remaining unredacted PII.
Materials and methods: Using 2000 representative clinical documents from 2 healthcare settings (4000 total), we used a novel method to generate 2 de-identified 100-document corpora (200 documents total) in which PII tagged by a typical automated machine-learned tagger was replaced by HIPS-resynthesized content. Four readers conducted aggressive reidentification attacks to isolate leaked PII: 2 readers from within the originating institution and 2 external readers.
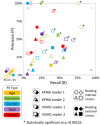
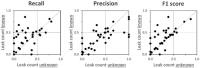
Results: Overall, mean recall of leaked PII was 26.8% and mean precision was 37.2%. Mean recall was 9% (mean precision = 37%) for patient ages, 32% (mean precision = 26%) for dates, 25% (mean precision = 37%) for doctor names, 45% (mean precision = 55%) for organization names, and 23% (mean precision = 57%) for patient names. Recall was 32% (precision = 40%) for internal and 22% (precision =33%) for external readers.
Discussion and conclusions: Approximately 70% of leaked PII "hiding" in a corpus de-identified with HIPS resynthesis is resilient to detection by human readers in a realistic, aggressive reidentification attack scenario-more than double the rate reported in previous studies but less than the rate reported for an attack assisted by machine learning methods.
Keywords: biomedical research; confidentiality; de-identification; electronic health records; natural language processing; privacy.
© The Author(s) 2020. Published by Oxford University Press on behalf of the American Medical Informatics Association. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures

Similar articles
-
The machine giveth and the machine taketh away: a parrot attack on clinical text deidentified with hiding in plain sight.J Am Med Inform Assoc. 2019 Dec 1;26(12):1536-1544. doi: 10.1093/jamia/ocz114. J Am Med Inform Assoc. 2019. PMID: 31390016 Free PMC article.
-
Hiding in plain sight: use of realistic surrogates to reduce exposure of protected health information in clinical text.J Am Med Inform Assoc. 2013 Mar-Apr;20(2):342-8. doi: 10.1136/amiajnl-2012-001034. Epub 2012 Jul 6. J Am Med Inform Assoc. 2013. PMID: 22771529 Free PMC article.
-
An Extensible Evaluation Framework Applied to Clinical Text Deidentification Natural Language Processing Tools: Multisystem and Multicorpus Study.J Med Internet Res. 2024 May 28;26:e55676. doi: 10.2196/55676. J Med Internet Res. 2024. PMID: 38805692 Free PMC article.
-
De-identification of free text data containing personal health information: a scoping review of reviews.Int J Popul Data Sci. 2023 Dec 12;8(1):2153. doi: 10.23889/ijpds.v8i1.2153. eCollection 2023. Int J Popul Data Sci. 2023. PMID: 38414537 Free PMC article.
-
Accuracy of privacy preserving record linkage for real world data in the United States: a systemic review.JAMIA Open. 2025 Jan 22;8(1):ooaf002. doi: 10.1093/jamiaopen/ooaf002. eCollection 2025 Feb. JAMIA Open. 2025. PMID: 39845287 Free PMC article. Review.
Cited by
-
Informatics impact requires effective, scalable tools and standards-based infrastructure.J Am Med Inform Assoc. 2020 Jul 1;27(9):1341-1342. doi: 10.1093/jamia/ocaa187. J Am Med Inform Assoc. 2020. PMID: 32989458 Free PMC article. No abstract available.

