Bacteria-related changes in host DNA methylation and the risk for CRC
- PMID: 32931352
- PMCID: PMC7575230
- DOI: 10.1080/19490976.2020.1800898
Bacteria-related changes in host DNA methylation and the risk for CRC
Abstract
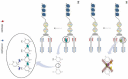
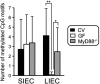
Colorectal cancer (CRC) is the second most common cause of cancer deaths in men and women combined. Colon-tumor growth is multistage and the result of the accumulation of spontaneous mutations and epigenetic events that silence tumor-suppressor genes and activate oncogenes. Environmental factors are primary contributors to these somatic gene alterations, which account for the increase in incidence of CRC in western countries. In recent decades, gut microbiota and their metabolites have been recognized as essential contributing factors to CRC, and now serve as biomarkers for the diagnosis and prognosis of CRC. In the present review, we highlight holistic approaches to understanding how gut microbiota contributes to CRC. We particularly focus herein on bacteria-related changes in host DNA methylation and the risk for CRC.
Keywords: Colon; cancer; methylation; microbiota; mutation.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
