CoV-Seq, a New Tool for SARS-CoV-2 Genome Analysis and Visualization: Development and Usability Study
- PMID: 32931441
- PMCID: PMC7537720
- DOI: 10.2196/22299
CoV-Seq, a New Tool for SARS-CoV-2 Genome Analysis and Visualization: Development and Usability Study
Abstract
Background: COVID-19 became a global pandemic not long after its identification in late 2019. The genomes of SARS-CoV-2 are being rapidly sequenced and shared on public repositories. To keep up with these updates, scientists need to frequently refresh and reclean data sets, which is an ad hoc and labor-intensive process. Further, scientists with limited bioinformatics or programming knowledge may find it difficult to analyze SARS-CoV-2 genomes.
Objective: To address these challenges, we developed CoV-Seq, an integrated web server that enables simple and rapid analysis of SARS-CoV-2 genomes.
Methods: CoV-Seq is implemented in Python and JavaScript. The web server and source code URLs are provided in this article.
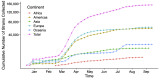
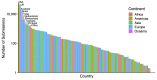
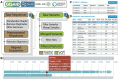
Results: Given a new sequence, CoV-Seq automatically predicts gene boundaries and identifies genetic variants, which are displayed in an interactive genome visualizer and are downloadable for further analysis. A command-line interface is available for high-throughput processing. In addition, we aggregated all publicly available SARS-CoV-2 sequences from the Global Initiative on Sharing Avian Influenza Data (GISAID), National Center for Biotechnology Information (NCBI), European Nucleotide Archive (ENA), and China National GeneBank (CNGB), and extracted genetic variants from these sequences for download and downstream analysis. The CoV-Seq database is updated weekly.
Conclusions: We have developed CoV-Seq, an integrated web service for fast and easy analysis of custom SARS-CoV-2 sequences. The web server provides an interactive module for the analysis of custom sequences and a weekly updated database of genetic variants of all publicly accessible SARS-CoV-2 sequences. We believe CoV-Seq will help improve our understanding of the genetic underpinnings of COVID-19.
Keywords: COVID-19; SARS-CoV-2; bioinformatics; data sets; genetics; genome; programming; sequence; virus; web server.
©Boxiang Liu, Kaibo Liu, He Zhang, Liang Zhang, Yuchen Bian, Liang Huang. Originally published in the Journal of Medical Internet Research (http://www.jmir.org), 02.10.2020.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures
References
-
- Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020 May;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. http://europepmc.org/abstract/MED/32087114 - DOI - PMC - PubMed
-
- Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018 Dec 01;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. http://europepmc.org/abstract/MED/29790939 - DOI - PMC - PubMed
-
- Brister JR, Ako-Adjei D, Bao Y, Blinkova O. NCBI viral genomes resource. Nucleic Acids Res. 2015 Jan;43(Database issue):D571–7. doi: 10.1093/nar/gku1207. http://europepmc.org/abstract/MED/25428358 - DOI - PMC - PubMed
-
- Kanz C, Aldebert P, Althorpe N, Baker W, Baldwin A, Bates K, Browne P, van den Broek A, Castro M, Cochrane G, Duggan K, Eberhardt R, Faruque N, Gamble J, Diez FG, Harte N, Kulikova T, Lin Q, Lombard V, Lopez R, Mancuso R, McHale M, Nardone F, Silventoinen V, Sobhany S, Stoehr P, Tuli MA, Tzouvara K, Vaughan R, Wu D, Zhu W, Apweiler R. The EMBL Nucleotide Sequence Database. Nucleic Acids Res. 2005 Jan 01;33(Database issue):D29–33. doi: 10.1093/nar/gki098. http://europepmc.org/abstract/MED/15608199 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

