Polyphenism of a Novel Trait Integrated Rapidly Evolving Genes into Ancestrally Plastic Networks
- PMID: 32931588
- PMCID: PMC7826178
- DOI: 10.1093/molbev/msaa235
Polyphenism of a Novel Trait Integrated Rapidly Evolving Genes into Ancestrally Plastic Networks
Abstract
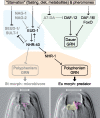
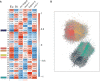
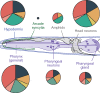
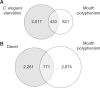
Developmental polyphenism, the ability to switch between phenotypes in response to environmental variation, involves the alternating activation of environmentally sensitive genes. Consequently, to understand how a polyphenic response evolves requires a comparative analysis of the components that make up environmentally sensitive networks. Here, we inferred coexpression networks for a morphological polyphenism, the feeding-structure dimorphism of the nematode Pristionchus pacificus. In this species, individuals produce alternative forms of a novel trait-moveable teeth, which in one morph enable predatory feeding-in response to environmental cues. To identify the origins of polyphenism network components, we independently inferred coexpression modules for more conserved transcriptional responses, including in an ancestrally nonpolyphenic nematode species. Further, through genome-wide analyses of these components across the nematode family (Diplogastridae) in which the polyphenism arose, we reconstructed how network components have changed. To achieve this, we assembled and resolved the phylogenetic context for five genomes of species representing the breadth of Diplogastridae and a hypothesized outgroup. We found that gene networks instructing alternative forms arose from ancestral plastic responses to environment, specifically starvation-induced metabolism and the formation of a conserved diapause (dauer) stage. Moreover, loci from rapidly evolving gene families were integrated into these networks with higher connectivity than throughout the rest of the P. pacificus transcriptome. In summary, we show that the modular regulatory outputs of a polyphenic response evolved through the integration of conserved plastic responses into networks with genes of high evolutionary turnover.
Keywords: coexpression network; developmental plasticity; modularity; nematodes; phylogenomics; taxon-restricted genes.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Abouheif E, Wray GA.. 2002. Evolution of the gene network underlying wing polyphenism in ants. Science 297(5579):249–252. - PubMed
-
- Antebi A, Culotti JG, Hedgecock EM.. 1998. daf-12 regulates developmental age and the dauer alternative in Caenorhabditis elegans. Development 125(7):1191–1205. - PubMed
-
- Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner PA.. 2016. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32(22):3380–3387. - PubMed
-
- Aubin-Horth N, Renn SCP.. 2009. Genomic reaction norms: using integrative biology to understand molecular mechanisms of phenotypic plasticity. Mol Ecol. 18(18):3763–3780. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

