The Expression Profile of mRNA and tRNA Genes in Splenocytes and Neutrophils after In Vivo Delivery of Antitumor Short Hairpin RNA of Indoleamine 2,3- Dioxygenase
- PMID: 32933162
- PMCID: PMC7555719
- DOI: 10.3390/ijms21186703
The Expression Profile of mRNA and tRNA Genes in Splenocytes and Neutrophils after In Vivo Delivery of Antitumor Short Hairpin RNA of Indoleamine 2,3- Dioxygenase
Abstract
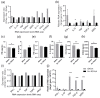
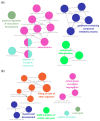
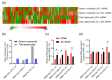
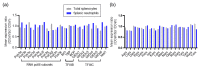
RNA-based therapeutics are considered as novel treatments for human diseases. Our previous study demonstrated that treatment with short-hairpin RNA against Ido1 (IDO shRNA) suppresses tumor growth, detects Th1-bias immune responses, and elevates expression of tryptophan transfer RNA (tRNATrp) in total splenocytes. In addition, depletion of Ly6g+ neutrophils attenuates the effect of IDO shRNA. The aim of this study was to investigate the regulatory network and the expression profile of tRNAs and other non-coding RNAs in IDO shRNA-treated spleens. The total splenocytes and magnetic bead-enriched splenic neutrophils were collected from the lung tumor bearing mice, which were treated with IDO shRNA or scramble IDO shRNA, and the collected cells were subsequently subjected to RNA sequencing. The gene ontology analysis revealed the different enrichment pathways in total splenocytes and splenic neutrophils. Furthermore, the expression of tRNA genes was identified and validated. Six isoacceptors of tRNA, with different expression patterns between total splenocytes and splenic neutrophils, were observed. In summary, our findings not only revealed novel biological processes in IDO shRNA-treated total splenocytes and splenic neutrophils, but the identified tRNAs and other non-coding RNAs may contribute to developing a novel biomarker gene set for evaluating the clinical efficiency of RNA-based cancer immunotherapies.
Keywords: RNA sequencing; animal tumor model; biomarker; indoleamine 2,3-dioxygenase1 (IDO1); neutrophil; non-coding RNA; short-hairpin RNA (shRNA); transfer RNA (tRNA).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- DeWeerdt S. RNA therapies explained. Nature. 2019;574:S2–S3. doi: 10.1038/d41586-019-03068-4. - DOI
MeSH terms
Substances
Grants and funding
- MOST 106-2320-B-037-029-MY3/Ministry of Science and Technology, Taiwan
- MOST 107-2320-B-037-011-MY3/Ministry of Science and Technology, Taiwan
- EDCHP-109013/E-DA Cancer Hospital
- KMUH107-7R81/Kaohsiung Medical University Chung-Ho Memorial Hospital
- KMUH108-8R72/Kaohsiung Medical University Chung-Ho Memorial Hospital
LinkOut - more resources
Full Text Sources
Research Materials

