Targeting SARS-CoV-2 Nsp12/Nsp8 interaction interface with approved and investigational drugs: an in silico structure-based approach
- PMID: 32933378
- PMCID: PMC7544933
- DOI: 10.1080/07391102.2020.1819882
Targeting SARS-CoV-2 Nsp12/Nsp8 interaction interface with approved and investigational drugs: an in silico structure-based approach
Abstract
In this study, the Nsp12-Nsp8 complex of SARS-CoV-2 was targeted with structure-based and computer-aided drug design approach because of its vital role in viral replication. Sequence analysis of RNA-dependent RNA polymerase (Nsp12) sequences from 30,366 different isolates were analysed for possible mutations. FDA-approved and investigational drugs were screened for interaction with both mutant and wild-type Nsp12-Nsp8 interfaces. Sequence analysis revealed that 70.42% of Nsp12 sequences showed conserved P323L mutation, located in the Nsp8 binding cleft. Compounds were screened for interface interaction, any with XP GScores lower than -7.0 kcal/mol were considered as possible interface inhibitors. RX-3117 (fluorocyclopentenyl cytosine) and Nebivolol had the highest binding affinities in both mutant and wild-type enzymes, therefore they were selected and resultant protein-ligand complexes were simulated for analysis of stability over 100 ns. Although the selected ligands had partial mobility in the binding cavity, they were not removed from the binding pocket after 100 ns. The ligand RX-3117 remained in the same position in the binding pocket of the mutant and wild-type enzyme after 100 ns MD simulation. However, the ligand Nebivolol folded and embedded in the binding pocket of mutant Nsp12 protein. Overall, FDA-approved and investigational drugs are able to bind to the Nsp12-Nsp8 interaction interface and prevent the formation of the Nsp12-Nsp8 complex. Interruption of viral replication by drugs proposed in this study should be further tested to pave the way for in vivo studies towards the treatment of COVID-19.Communicated by Ramaswamy H. Sarma.
Keywords: COVID-19; Nsp12; RNA-dependent RNA polymerase; SARS-CoV-2; drug repositioning; mutation analysis.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures


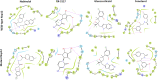

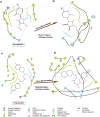
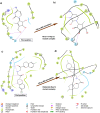

References
-
- Berendsen, H. J. C., Postma, J. P. M., Van Gunsteren, W. F., Dinola, A., & Haak, J. R. (1984). Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics, 81(8), 3684–3690. 10.1063/1.448118 - DOI
-
- Betts, M. J., & Russell, R. B. (2003). Amino acid properties and consequences of substitutions. In Barnes M. R. & Gray I. C. (Eds.), Bioinformatics for geneticists (pp. 289–316). Wiley. 10.1002/0470867302.ch14 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
