Biochemical and biophysical analyses of hypoxia sensing prolyl hydroxylases from Dictyostelium discoideum and Toxoplasma gondii
- PMID: 32934009
- PMCID: PMC7864055
- DOI: 10.1074/jbc.RA120.013998
Biochemical and biophysical analyses of hypoxia sensing prolyl hydroxylases from Dictyostelium discoideum and Toxoplasma gondii
Abstract
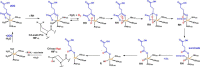
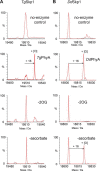
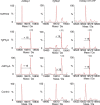
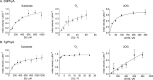
In animals, the response to chronic hypoxia is mediated by prolyl hydroxylases (PHDs) that regulate the levels of hypoxia-inducible transcription factor α (HIFα). PHD homologues exist in other types of eukaryotes and prokaryotes where they act on non HIF substrates. To gain insight into the factors underlying different PHD substrates and properties, we carried out biochemical and biophysical studies on PHD homologues from the cellular slime mold, Dictyostelium discoideum, and the protozoan parasite, Toxoplasma gondii, both lacking HIF. The respective prolyl-hydroxylases (DdPhyA and TgPhyA) catalyze prolyl-hydroxylation of S-phase kinase-associated protein 1 (Skp1), a reaction enabling adaptation to different dioxygen availability. Assays with full-length Skp1 substrates reveal substantial differences in the kinetic properties of DdPhyA and TgPhyA, both with respect to each other and compared with human PHD2; consistent with cellular studies, TgPhyA is more active at low dioxygen concentrations than DdPhyA. TgSkp1 is a DdPhyA substrate and DdSkp1 is a TgPhyA substrate. No cross-reactivity was detected between DdPhyA/TgPhyA substrates and human PHD2. The human Skp1 E147P variant is a DdPhyA and TgPhyA substrate, suggesting some retention of ancestral interactions. Crystallographic analysis of DdPhyA enables comparisons with homologues from humans, Trichoplax adhaerens, and prokaryotes, informing on differences in mobile elements involved in substrate binding and catalysis. In DdPhyA, two mobile loops that enclose substrates in the PHDs are conserved, but the C-terminal helix of the PHDs is strikingly absent. The combined results support the proposal that PHD homologues have evolved kinetic and structural features suited to their specific sensing roles.
Keywords: 2-oxoglutarate/α-ketoglutarate oxygenase; Dictyostelium discoideum; S-phase kinase-associated protein 1 (Skp1); Toxoplasma gondii; dioxygenase; hypoxia; hypoxia-inducible factor (HIF); hypoxia/oxygen sensor; prolyl-hydroxylase; protein evolution.
© 2020 Liu et al.
Conflict of interest statement
Conflict of interest—The authors declare no competing financial interests.
Figures



Similar articles
-
The Skp1 protein from Toxoplasma is modified by a cytoplasmic prolyl 4-hydroxylase associated with oxygen sensing in the social amoeba Dictyostelium.J Biol Chem. 2012 Jul 20;287(30):25098-110. doi: 10.1074/jbc.M112.355446. Epub 2012 May 30. J Biol Chem. 2012. PMID: 22648409 Free PMC article.
-
The E3 Ubiquitin Ligase Adaptor Protein Skp1 Is Glycosylated by an Evolutionarily Conserved Pathway That Regulates Protist Growth and Development.J Biol Chem. 2016 Feb 26;291(9):4268-80. doi: 10.1074/jbc.M115.703751. Epub 2015 Dec 30. J Biol Chem. 2016. PMID: 26719340 Free PMC article.
-
Kinetic Investigations of the Role of Factor Inhibiting Hypoxia-inducible Factor (FIH) as an Oxygen Sensor.J Biol Chem. 2015 Aug 7;290(32):19726-42. doi: 10.1074/jbc.M115.653014. Epub 2015 Jun 25. J Biol Chem. 2015. PMID: 26112411 Free PMC article.
-
Enzyme substrate recognition in oxygen sensing: how the HIF trap snaps.Biochem J. 2007 Dec 1;408(2):e5-6. doi: 10.1042/BJ20071306. Biochem J. 2007. PMID: 17990984 Free PMC article. Review.
-
Protein Hydroxylation by Hypoxia-Inducible Factor (HIF) Hydroxylases: Unique or Ubiquitous?Cells. 2019 Apr 26;8(5):384. doi: 10.3390/cells8050384. Cells. 2019. PMID: 31035491 Free PMC article. Review.
Cited by
-
Gas Tunnel Engineering of Prolyl Hydroxylase Reprograms Hypoxia Signaling in Cells.Angew Chem Int Ed Engl. 2024 Nov 25;63(48):e202409234. doi: 10.1002/anie.202409234. Epub 2024 Oct 21. Angew Chem Int Ed Engl. 2024. PMID: 39168829
-
Glycosylation Weakens Skp1 Homodimerization in Toxoplasma gondii by Interrupting a Fuzzy Interaction.Biochemistry. 2025 May 20;64(10):2262-2279. doi: 10.1021/acs.biochem.4c00859. Epub 2025 Apr 29. Biochemistry. 2025. PMID: 40296701 Free PMC article.
-
Toxoplasma gondii harbors a hypoxia-responsive coproporphyrinogen dehydrogenase-like protein.bioRxiv [Preprint]. 2023 Nov 16:2023.11.16.567449. doi: 10.1101/2023.11.16.567449. bioRxiv. 2023. Update in: mSphere. 2024 Mar 26;9(3):e0009224. doi: 10.1128/msphere.00092-24. PMID: 38014006 Free PMC article. Updated. Preprint.
-
Toxoplasma gondii harbors a hypoxia-responsive coproporphyrinogen dehydrogenase-like protein.mSphere. 2024 Mar 26;9(3):e0009224. doi: 10.1128/msphere.00092-24. Epub 2024 Feb 27. mSphere. 2024. PMID: 38411121 Free PMC article.
-
Water deprivation-induced hypoxia and oxidative stress physiology responses in respiratory organs of the Indian stinging fish in near coastal zones.PeerJ. 2024 Jan 25;12:e16793. doi: 10.7717/peerj.16793. eCollection 2024. PeerJ. 2024. PMID: 38282857 Free PMC article.
References
-
- Jaakkola P., Mole D.R., Tian Y.M., Wilson M.I., Gielbert J., Gaskell S.J., von Kriegsheim A., Hebestreit H.F., Mukherji M., Schofield C.J., Maxwell P.H., Pugh C.W., Ratcliffe P.J. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. 11292861. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

