New insights into non-transcriptional regulation of mammalian core clock proteins
- PMID: 32934011
- PMCID: PMC7520459
- DOI: 10.1242/jcs.241174
New insights into non-transcriptional regulation of mammalian core clock proteins
Abstract
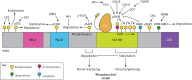
Mammalian circadian rhythms drive ∼24 h periodicity in a wide range of cellular processes, temporally coordinating physiology and behaviour within an organism, and synchronising this with the external day-night cycle. The canonical model for this timekeeping consists of a delayed negative-feedback loop, containing transcriptional activator complex CLOCK-BMAL1 (BMAL1 is also known as ARNTL) and repressors period 1, 2 and 3 (PER1, PER2 and PER3) and cryptochrome 1 and 2 (CRY1 and CRY2), along with a number of accessory factors. Although the broad strokes of this system are defined, the exact molecular mechanisms by which these proteins generate a self-sustained rhythm with such periodicity and fidelity remains a topic of much research. Recent studies have identified prominent roles for a number of crucial post-transcriptional, translational and, particularly, post-translational events within the mammalian circadian oscillator, providing an increasingly complex understanding of the activities and interactions of the core clock proteins. In this Review, we highlight such contemporary work on non-transcriptional events and set it within our current understanding of cellular circadian timekeeping.
Keywords: Cellular timekeeping; Circadian rhythm; Post-transcriptional modification; Post-translational modification; Translational regulation.
© 2020. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures


References
-
- Atger F., Gobet C., Marquis J., Martin E., Wang J., Weger B., Lefebvre G., Descombes P., Naef F. and Gachon F. (2015). Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc. Natl. Acad. Sci. USA 112, E6579-E6588. 10.1073/pnas.1515308112 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

