Offloading Role of a Discrete Thioesterase in Type II Polyketide Biosynthesis
- PMID: 32934080
- PMCID: PMC7492732
- DOI: 10.1128/mBio.01334-20
Offloading Role of a Discrete Thioesterase in Type II Polyketide Biosynthesis
Abstract
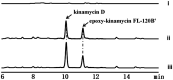
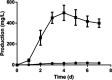
Type II polyketides are a group of secondary metabolites with various biological activities. In nature, biosynthesis of type II polyketides involves multiple enzymatic steps whereby key enzymes, including ketoacyl-synthase (KSα), chain length factor (KSβ), and acyl carrier protein (ACP), are utilized to elongate the polyketide chain through a repetitive condensation reaction. During each condensation, the biosynthesis intermediates are covalently attached to KSα or ACP via a thioester bond and are then cleaved to release an elongated polyketide chain for successive postmodification. Despite its critical role in type II polyketide biosynthesis, the enzyme and its corresponding mechanism for type II polyketide chain release through thioester bond breakage have yet to be determined. Here, kinamycin was used as a model compound to investigate the chain release step of type II polyketide biosynthesis. Using a genetic knockout strategy, we confirmed that AlpS is required for the complete biosynthesis of kinamycins. Further in vitro biochemical assays revealed high hydrolytic activity of AlpS toward a thioester bond in an aromatic polyketide-ACP analog, suggesting its distinct role in offloading the polyketide chain from ACP during the kinamycin biosynthesis. Finally, we successfully utilized AlpS to enhance the heterologous production of dehydrorabelomycin in Escherichia coli by nearly 25-fold, which resulted in 0.50 g/liter dehydrorabelomycin in a simple batch-mode shake flask culture. Taken together, our results provide critical knowledge to gain an insightful understanding of the chain-releasing process during type II polyketide synthesis, which, in turn, lays a solid foundation for future new applications in type II polyketide bioproduction.
Keywords: biosynthesis; natural product; offloading; thioesterase; type II polyketides.
Copyright © 2020 Hua et al.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

