Mechanistic Understanding Enables the Rational Design of Salicylanilide Combination Therapies for Gram-Negative Infections
- PMID: 32934086
- PMCID: PMC7492738
- DOI: 10.1128/mBio.02068-20
Mechanistic Understanding Enables the Rational Design of Salicylanilide Combination Therapies for Gram-Negative Infections
Abstract
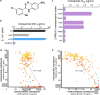
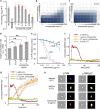
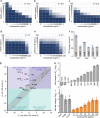
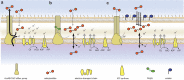
One avenue to combat multidrug-resistant Gram-negative bacteria is the coadministration of multiple drugs (combination therapy), which can be particularly promising if drugs synergize. The identification of synergistic drug combinations, however, is challenging. Detailed understanding of antibiotic mechanisms can address this issue by facilitating the rational design of improved combination therapies. Here, using diverse biochemical and genetic assays, we examine the molecular mechanisms of niclosamide, a clinically approved salicylanilide compound, and demonstrate its potential for Gram-negative combination therapies. We discovered that Gram-negative bacteria possess two innate resistance mechanisms that reduce their niclosamide susceptibility: a primary mechanism mediated by multidrug efflux pumps and a secondary mechanism of nitroreduction. When efflux was compromised, niclosamide became a potent antibiotic, dissipating the proton motive force (PMF), increasing oxidative stress, and reducing ATP production to cause cell death. These insights guided the identification of diverse compounds that synergized with salicylanilides when coadministered (efflux inhibitors, membrane permeabilizers, and antibiotics that are expelled by PMF-dependent efflux), thus suggesting that salicylanilide compounds may have broad utility in combination therapies. We validate these findings in vivo using a murine abscess model, where we show that niclosamide synergizes with the membrane permeabilizing antibiotic colistin against high-density infections of multidrug-resistant Gram-negative clinical isolates. We further demonstrate that enhanced nitroreductase activity is a potential route to adaptive niclosamide resistance but show that this causes collateral susceptibility to clinical nitro-prodrug antibiotics. Thus, we highlight how mechanistic understanding of mode of action, innate/adaptive resistance, and synergy can rationally guide the discovery, development, and stewardship of novel combination therapies.IMPORTANCE There is a critical need for more-effective treatments to combat multidrug-resistant Gram-negative infections. Combination therapies are a promising strategy, especially when these enable existing clinical drugs to be repurposed as antibiotics. We examined the mechanisms of action and basis of innate Gram-negative resistance for the anthelmintic drug niclosamide and subsequently exploited this information to demonstrate that niclosamide and analogs kill Gram-negative bacteria when combined with antibiotics that inhibit drug efflux or permeabilize membranes. We confirm the synergistic potential of niclosamide in vitro against a diverse range of recalcitrant Gram-negative clinical isolates and in vivo in a mouse abscess model. We also demonstrate that nitroreductases can confer resistance to niclosamide but show that evolution of these enzymes for enhanced niclosamide resistance confers a collateral sensitivity to other clinical antibiotics. Our results highlight how detailed mechanistic understanding can accelerate the evaluation and implementation of new combination therapies.
Keywords: antibiotic resistance; colistin; drug efflux; efflux; niclosamide; nitroreductase; proton motive force; repurposing; resistance; synergy.
Copyright © 2020 Copp et al.
Figures
Similar articles
-
The Anthelmintic Drug Niclosamide Synergizes with Colistin and Reverses Colistin Resistance in Gram-Negative Bacilli.Antimicrob Agents Chemother. 2019 Mar 27;63(4):e02574-18. doi: 10.1128/AAC.02574-18. Print 2019 Apr. Antimicrob Agents Chemother. 2019. PMID: 30917988 Free PMC article.
-
Synergistic combinations of anthelmintic salicylanilides oxyclozanide, rafoxanide, and closantel with colistin eradicates multidrug-resistant colistin-resistant Gram-negative bacilli.J Antibiot (Tokyo). 2019 Aug;72(8):605-616. doi: 10.1038/s41429-019-0186-8. Epub 2019 Apr 26. J Antibiot (Tokyo). 2019. PMID: 31028351
-
Repurposing salicylanilide anthelmintic drugs to combat drug resistant Staphylococcus aureus.PLoS One. 2015 Apr 21;10(4):e0124595. doi: 10.1371/journal.pone.0124595. eCollection 2015. PLoS One. 2015. PMID: 25897961 Free PMC article.
-
Efflux-mediated Multidrug Resistance in Critical Gram-negative Bacteria and Natural Efflux Pump Inhibitors.Curr Drug Res Rev. 2024;16(3):349-368. doi: 10.2174/0125899775271214240112071830. Curr Drug Res Rev. 2024. PMID: 38288795 Review.
-
Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria.FEMS Microbiol Rev. 2012 Mar;36(2):340-63. doi: 10.1111/j.1574-6976.2011.00290.x. Epub 2011 Jul 29. FEMS Microbiol Rev. 2012. PMID: 21707670 Free PMC article. Review.
Cited by
-
Enhanced Antibacterial Activity of Repurposed Mitomycin C and Imipenem in Combination with the Lytic Phage vB_KpnM-VAC13 against Clinical Isolates of Klebsiella pneumoniae.Antimicrob Agents Chemother. 2021 Aug 17;65(9):e0090021. doi: 10.1128/AAC.00900-21. Epub 2021 Aug 17. Antimicrob Agents Chemother. 2021. PMID: 34228538 Free PMC article.
-
Amphiphilic mPEG-PLGA copolymer nanoparticles co-delivering colistin and niclosamide to treat colistin-resistant Gram-negative bacteria infections.Commun Biol. 2025 Apr 29;8(1):673. doi: 10.1038/s42003-025-08095-8. Commun Biol. 2025. PMID: 40295783 Free PMC article.
-
Study of Biological Activities and ADMET-Related Properties of Salicylanilide-Based Peptidomimetics.Int J Mol Sci. 2022 Oct 1;23(19):11648. doi: 10.3390/ijms231911648. Int J Mol Sci. 2022. PMID: 36232947 Free PMC article.
-
The chemotherapeutic drug methotrexate selects for antibiotic resistance.EBioMedicine. 2021 Dec;74:103742. doi: 10.1016/j.ebiom.2021.103742. Epub 2021 Dec 11. EBioMedicine. 2021. PMID: 34902789 Free PMC article.
-
Cationic Polymers Enable Internalization of Negatively Charged Chemical Probes into Bacteria.ACS Chem Biol. 2023 Sep 15;18(9):2063-2072. doi: 10.1021/acschembio.3c00351. Epub 2023 Sep 6. ACS Chem Biol. 2023. PMID: 37671702 Free PMC article.
References
-
- World Health Organization. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. WHO, Geneva, Switzerland.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

