Evidence That VirS Is a Receptor for the Signaling Peptide of the Clostridium perfringens Agr-like Quorum Sensing System
- PMID: 32934089
- PMCID: PMC7492741
- DOI: 10.1128/mBio.02219-20
Evidence That VirS Is a Receptor for the Signaling Peptide of the Clostridium perfringens Agr-like Quorum Sensing System
Abstract
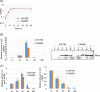
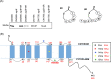
Since both the Agr (accessory gene regulator)-like quorum sensing (QS) system and VirS/VirR (VirS/R) two-component regulatory system of Clostridium perfringens positively regulate production of several toxins, including C. perfringens beta toxin (CPB), it has been hypothesized the VirS membrane sensor protein is an Agr-like QS signaling peptide (SP) receptor. To begin evaluating whether VirS is an SP receptor, this study sequenced the virS gene in C. perfringens strains CN3685 and CN1795 because it was reported that agrB mutants of both strains increase CPB production in response to the pentapeptide 5R, likely the natural SP, but only the CN3685 agrB mutant responds to 8R, which is 5R plus a 3-amino-acid tail. This sequencing identified differences between the predicted VirS extracellular loop 2 (ECL2) of CN3685 versus that of CN1795. To explore if those ECL2 differences explain strain-related variations in SP sensitivity and support VirS as an SP receptor, virS agrB double-null mutants of each strain were complemented to swap which VirS protein they produce. CPB Western blotting showed that this complementation changed the natural responsiveness of each strain to 8R. A pulldown experiment using biotin-5R demonstrated that VirS can bind SP. To further support VirS:SP binding and to identify a VirS binding site for SP, a 14-mer peptide corresponding to VirS ECL2 was synthesized. This ECL2 peptide inhibited 5R signaling to agrB mutant and wild-type strains. This inhibition was specific, since a single N to D substitution in the ECL2 peptide abrogated these effects. Collectively, these results support VirS as an important SP receptor and may assist development of therapeutics.IMPORTANCEC. perfringens beta toxin (CPB) is essential for the virulence of type C strains, a common cause of fatal necrotizing enteritis and enterotoxemia in humans and domestic animals. Production of CPB, as well as several other C. perfringens toxins, is positively regulated by both the Agr-like QS system and the VirS/R two-component regulatory system. This study presents evidence that the VirS membrane sensor protein is a receptor for the AgrD-derived SP and that the second extracellular loop of VirS is important for SP binding. Understanding interactions between SP and VirS improves knowledge of C. perfringens pathogenicity and may provide insights for designing novel strategies to reduce C. perfringens toxin production during infections.
Keywords: Agr-like quorum sensing; Clostridium perfringens; VirS/R two-component regulatory system; beta toxin; signal peptide receptor; toxin production regulation.
Copyright © 2020 Li and McClane.
Figures


Similar articles
-
Identifying the Basis for VirS/VirR Two-Component Regulatory System Control of Clostridium perfringens Beta-Toxin Production.J Bacteriol. 2021 Aug 20;203(18):e0027921. doi: 10.1128/JB.00279-21. Epub 2021 Aug 20. J Bacteriol. 2021. PMID: 34228498 Free PMC article.
-
Epsilon-toxin production by Clostridium perfringens type D strain CN3718 is dependent upon the agr operon but not the VirS/VirR two-component regulatory system.mBio. 2011 Dec 13;2(6):e00275-11. doi: 10.1128/mBio.00275-11. Print 2011. mBio. 2011. Retraction in: mBio. 2022 Apr 26;13(2):e0049522. doi: 10.1128/mbio.00495-22. PMID: 22167225 Free PMC article. Retracted.
-
Structure-function analysis of peptide signaling in the Clostridium perfringens Agr-like quorum sensing system.J Bacteriol. 2015 May;197(10):1807-18. doi: 10.1128/JB.02614-14. Epub 2015 Mar 16. J Bacteriol. 2015. PMID: 25777675 Free PMC article.
-
Host cell-induced signaling causes Clostridium perfringens to upregulate production of toxins important for intestinal infections.Gut Microbes. 2014 Jan-Feb;5(1):96-107. doi: 10.4161/gmic.26419. Epub 2013 Sep 10. Gut Microbes. 2014. PMID: 24061146 Free PMC article. Review.
-
Gene regulation by the VirS/VirR system in Clostridium perfringens.Anaerobe. 2016 Oct;41:5-9. doi: 10.1016/j.anaerobe.2016.06.003. Epub 2016 Jun 11. Anaerobe. 2016. PMID: 27296833 Review.
Cited by
-
Autoinducing peptide-based quorum signaling systems in Clostridioides difficile.Curr Opin Microbiol. 2022 Feb;65:81-86. doi: 10.1016/j.mib.2021.10.017. Epub 2021 Nov 10. Curr Opin Microbiol. 2022. PMID: 34773906 Free PMC article. Review.
-
Identification of orphan histidine kinases that impact sporulation and enterotoxin production by Clostridium perfringens type F strain SM101 in a pathophysiologically-relevant ex vivo mouse intestinal contents model.PLoS Pathog. 2023 Jun 1;19(6):e1011429. doi: 10.1371/journal.ppat.1011429. eCollection 2023 Jun. PLoS Pathog. 2023. PMID: 37262083 Free PMC article.
-
Identifying the Basis for VirS/VirR Two-Component Regulatory System Control of Clostridium perfringens Beta-Toxin Production.J Bacteriol. 2021 Aug 20;203(18):e0027921. doi: 10.1128/JB.00279-21. Epub 2021 Aug 20. J Bacteriol. 2021. PMID: 34228498 Free PMC article.
-
Anemoside B4 attenuates necrotic enteritis of laying hens induced by Clostridium perfringens via inhibiting NF-κB and PI3K/Akt/mTOR signalling pathways.Heliyon. 2024 Jun 15;10(12):e33161. doi: 10.1016/j.heliyon.2024.e33161. eCollection 2024 Jun 30. Heliyon. 2024. PMID: 39005924 Free PMC article.
-
Pathogenicity and virulence of Clostridium perfringens.Virulence. 2021 Dec;12(1):723-753. doi: 10.1080/21505594.2021.1886777. Virulence. 2021. PMID: 33843463 Free PMC article.
References
-
- McClane BA, Uzal FA, Mf M, D L, Td W. 2006. The enterotoxic clostridia, p 688–752. In Dworkin M, Falkow S, Rosenburg E, Schleifer H, Stackebrandt E (ed), The prokaryotes, 3rd ed Springer Press, New York, NY.
-
- Rood JI. 2006. Clostridium perfringens and histotoxic disease, p 753–770. In Dworkin M, Falkow S, Rosenburg E, Schleifer H, Stackebrandt E (ed), The prokaryotes, 3rd ed Springer Press, New York, NY.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

