Influence of host genetics in shaping the rumen bacterial community in beef cattle
- PMID: 32934296
- PMCID: PMC7493918
- DOI: 10.1038/s41598-020-72011-9
Influence of host genetics in shaping the rumen bacterial community in beef cattle
Abstract
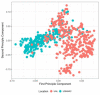
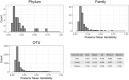
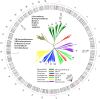
In light of recent host-microbial association studies, a consensus is evolving that species composition of the gastrointestinal microbiota is a polygenic trait governed by interactions between host genetic factors and the environment. Here, we investigated the effect of host genetic factors in shaping the bacterial species composition in the rumen by performing a genome-wide association study. Using a common set of 61,974 single-nucleotide polymorphisms found in cattle genomes (n = 586) and corresponding rumen bacterial community composition, we identified operational taxonomic units (OTUs), Families and Phyla with high heritability. The top associations (1-Mb windows) were located on 7 chromosomes. These regions were associated with the rumen microbiota in multiple ways; some (chromosome 19; position 3.0-4.0 Mb) are associated with closely related taxa (Prevotellaceae, Paraprevotellaceae, and RF16), some (chromosome 27; position 3.0-4.0 Mb) are associated with distantly related taxa (Prevotellaceae, Fibrobacteraceae, RF16, RFP12, S24-7, Lentisphaerae, and Tenericutes) and others (chromosome 23; position 0.0-1.0) associated with both related and unrelated taxa. The annotated genes associated with identified genomic regions suggest the associations observed are directed toward selective absorption of volatile fatty acids from the rumen to increase energy availability to the host. This study demonstrates that host genetics affects rumen bacterial community composition.
Conflict of interest statement
Samodha C. Fernando, author of this publication has disclosed a significant financial interest in NuGUT LLC. In accordance with its Conflict of Interest policy, the University of Nebraska-Lincoln’s Conflict of Interest in Research Committee has determined that this must be disclosed. The rest of the authors have nothing to disclose.
Figures



References
-
- Church DC. The Ruminant Animal Digestive Physiology and Nutrition. Long Grove: Waveland Press Inc; 1993.
-
- Hobson PN. The Rumen Microbial Ecosystem. London: Elsevier Applied Science; 1988.
-
- Hungate RE. The Rumen and Its Microbes. London: Academic Press Inc; 1966.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

