Maternal embryonic leucine zipper kinase: A novel biomarker and a potential therapeutic target in lung adenocarcinoma
- PMID: 32934715
- PMCID: PMC7471708
- DOI: 10.3892/ol.2020.12010
Maternal embryonic leucine zipper kinase: A novel biomarker and a potential therapeutic target in lung adenocarcinoma
Abstract
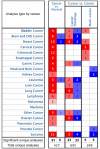
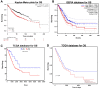
Maternal embryonic leucine zipper kinase (MELK), is an adenosine monophosphate-activated protein kinase-related kinase that serves important roles in tumourigenesis in multiple malignant tumours. However, to the best of our knowledge, the effect of MELK in lung adenocarcinoma (LUAD) has not been elucidated. The present study aimed to explore the clinical significance of MELK in the prognosis of LUAD. Data from Oncomine, Gene Expression Profiling Interactive Analysis (GEPIA) and The Cancer Genome Atlas (TCGA) were selected to predict the differential mRNA expression levels of MELK mRNA in LUAD and normal tissues. Subsequently, LUAD and adjacent normal tissue samples were collected from 75 patients with the disease, and immunohistochemistry was used to detect the protein expression of MELK. In addition, the Kaplan-Meier Plotter database, GEPIA and TCGA were used to verify the effect of MELK expression on clinical prognosis in patients with LUAD. MELK was significantly upregulated in LUAD tissues compared with that in normal tissues based on Oncomine, GEPIA and TCGA data (P<0.05). In addition, the results from immunohistochemistry demonstrated that the MELK protein level in LUAD tissues was significantly higher compared with that in matched normal tissues (P<0.05). Prognostic analysis performed using the Kaplan-Meier plotter, GEPIA and TCGA suggested that the expression of MELK was negatively associated with the overall survival time of patients with LUAD (P<0.05). In conclusion, MELK was highly expressed in LUAD based on bioinformatics and immunohistochemistry analysis, and increased expression of MELK was associated with a poor patient prognosis. MELK may serve as a potential diagnostic marker and therapeutic target for LUAD.
Keywords: MELK; bioinformatics; immuno-histochemistry; lung adenocarcinoma; prognosis.
Copyright: © Chen et al.
Figures



Similar articles
-
Decoding the significant diagnostic and prognostic importance of maternal embryonic leucine zipper kinase in human cancers through deep integrative analyses.J Cancer Res Ther. 2023 Oct 1;19(7):1852-1864. doi: 10.4103/jcrt.jcrt_1902_21. Epub 2023 Dec 29. J Cancer Res Ther. 2023. PMID: 38376289
-
Elevated Heterogeneous Nuclear Ribonucleoprotein C Expression Correlates With Poor Prognosis in Patients With Surgically Resected Lung Adenocarcinoma.Front Oncol. 2021 Jan 25;10:598437. doi: 10.3389/fonc.2020.598437. eCollection 2020. Front Oncol. 2021. PMID: 33569346 Free PMC article.
-
Up-Regulation of MELK Promotes Cell Growth and Invasion by Accelerating G1/S Transition and Indicates Poor Prognosis in Lung Adenocarcinoma.Mol Biotechnol. 2025 Apr;67(4):1584-1596. doi: 10.1007/s12033-024-01143-4. Epub 2024 Apr 27. Mol Biotechnol. 2025. PMID: 38676754
-
Analysis of the Expression of Cell Division Cycle-Associated Genes and Its Prognostic Significance in Human Lung Carcinoma: A Review of the Literature Databases.Biomed Res Int. 2020 Feb 12;2020:6412593. doi: 10.1155/2020/6412593. eCollection 2020. Biomed Res Int. 2020. PMID: 32104702 Free PMC article. Review.
-
Maternal embryonic leucine zipper kinase in tumor cells and tumor microenvironment: An emerging player and promising therapeutic opportunity.Cancer Lett. 2023 Apr 28;560:216126. doi: 10.1016/j.canlet.2023.216126. Epub 2023 Mar 16. Cancer Lett. 2023. PMID: 36933780 Review.
Cited by
-
MELK aggravates lung adenocarcinoma by regulating EZH2 ubiquitination and H3K27me3 histone methylation of LATS2.J Cell Mol Med. 2024 Apr;28(8):e18216. doi: 10.1111/jcmm.18216. J Cell Mol Med. 2024. PMID: 38652219 Free PMC article.
-
The high expression of TOP2A and MELK induces the occurrence of psoriasis.Aging (Albany NY). 2024 Feb 20;16(4):3185-3199. doi: 10.18632/aging.205519. Epub 2024 Feb 20. Aging (Albany NY). 2024. PMID: 38382096 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous
