This is a preprint.
Identifying zoonotic origin of SARS-CoV-2 by modeling the binding affinity between Spike receptor-binding domain and host ACE2
- PMID: 32935105
- PMCID: PMC7491519
- DOI: 10.1101/2020.09.11.293449
Identifying zoonotic origin of SARS-CoV-2 by modeling the binding affinity between Spike receptor-binding domain and host ACE2
Update in
-
Identifying the Zoonotic Origin of SARS-CoV-2 by Modeling the Binding Affinity between the Spike Receptor-Binding Domain and Host ACE2.J Proteome Res. 2020 Dec 4;19(12):4844-4856. doi: 10.1021/acs.jproteome.0c00717. Epub 2020 Nov 11. J Proteome Res. 2020. PMID: 33175551 Free PMC article.
Abstract
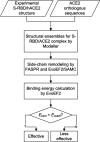
Despite considerable research progress on SARS-CoV-2, the direct zoonotic origin (intermediate host) of the virus remains ambiguous. The most definitive approach to identify the intermediate host would be the detection of SARS-CoV-2-like coronaviruses in wild animals. However, due to the high number of animal species, it is not feasible to screen all the species in the laboratory. Given that the recognition of the binding ACE2 proteins is the first step for the coronaviruses to invade host cells, we proposed a computational pipeline to identify potential intermediate hosts of SARS-CoV-2 by modeling the binding affinity between the Spike receptor-binding domain (RBD) and host ACE2. Using this pipeline, we systematically examined 285 ACE2 variants from mammals, birds, fish, reptiles, and amphibians, and found that the binding energies calculated on the modeled Spike-RBD/ACE2 complex structures correlate closely with the effectiveness of animal infections as determined by multiple experimental datasets. Built on the optimized binding affinity cutoff, we suggested a set of 96 mammals, including 48 experimentally investigated ones, which are permissive to SARS-CoV-2, with candidates from primates, rodents, and carnivores at the highest risk of infection. Overall, this work not only suggested a limited range of potential intermediate SARS-CoV-2 hosts for further experimental investigation; but more importantly, it proposed a new structure-based approach to general zoonotic origin and susceptibility analyses that are critical for human infectious disease control and wildlife protection.
Conflict of interest statement
Conflicts of interest
The authors declare that no conflict interest exists.
Figures



Similar articles
-
Identifying the Zoonotic Origin of SARS-CoV-2 by Modeling the Binding Affinity between the Spike Receptor-Binding Domain and Host ACE2.J Proteome Res. 2020 Dec 4;19(12):4844-4856. doi: 10.1021/acs.jproteome.0c00717. Epub 2020 Nov 11. J Proteome Res. 2020. PMID: 33175551 Free PMC article.
-
Computational modeling of the effect of five mutations on the structure of the ACE2 receptor and their correlation with infectivity and virulence of some emerged variants of SARS-CoV-2 suggests mechanisms of binding affinity dysregulation.Chem Biol Interact. 2022 Dec 1;368:110244. doi: 10.1016/j.cbi.2022.110244. Epub 2022 Nov 3. Chem Biol Interact. 2022. PMID: 36336003 Free PMC article.
-
Evaluating the transmission feasibility of SARS-CoV-2 Omicron (B.1.1.529) variant to 143 mammalian hosts: insights from S protein RBD and host ACE2 interaction studies.Funct Integr Genomics. 2023 Jan 12;23(1):36. doi: 10.1007/s10142-023-00962-z. Funct Integr Genomics. 2023. PMID: 36631570 Free PMC article.
-
Assessing the SARS-CoV-2 threat to wildlife: Potential risk to a broad range of mammals.Perspect Ecol Conserv. 2020 Oct-Dec;18(4):223-234. doi: 10.1016/j.pecon.2020.09.008. Epub 2020 Oct 5. Perspect Ecol Conserv. 2020. PMID: 33043253 Free PMC article. Review.
-
Interactions of angiotensin-converting enzyme-2 (ACE2) and SARS-CoV-2 spike receptor-binding domain (RBD): a structural perspective.Mol Biol Rep. 2023 Mar;50(3):2713-2721. doi: 10.1007/s11033-022-08193-4. Epub 2022 Dec 23. Mol Biol Rep. 2023. PMID: 36562937 Free PMC article. Review.
References
-
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020; 579:270–273. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
