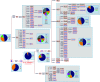
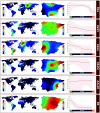
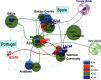
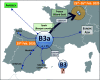
Phylogeography of SARS-CoV-2 pandemic in Spain: a story of multiple introductions, micro-geographic stratification, founder effects, and super-spreaders
- PMID: 32935498
- PMCID: PMC7671907
- DOI: 10.24272/j.issn.2095-8137.2020.217
Phylogeography of SARS-CoV-2 pandemic in Spain: a story of multiple introductions, micro-geographic stratification, founder effects, and super-spreaders
Abstract
Spain has been one of the main global pandemic epicenters for coronavirus disease 2019 (COVID-19). Here, we analyzed >41 000 genomes (including >26 000 high-quality (HQ) genomes) downloaded from the GISAID repository, including 1 245 (922 HQ) sampled in Spain. The aim of this study was to investigate genome variation of novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and reconstruct phylogeographic and transmission patterns in Spain. Phylogeographic analysis suggested at least 34 independent introductions of SARS-CoV-2 to Spain at the beginning of the outbreak. Six lineages spread very successfully in the country, probably favored by super-spreaders, namely, A2a4 (7.8%), A2a5 (38.4%), A2a10 (2.8%), B3a (30.1%), and B9 (8.7%), which accounted for 87.9% of all genomes in the Spanish database. One distinct feature of the Spanish SARS-CoV-2 genomes was the higher frequency of B lineages (39.3%, mainly B3a+B9) than found in any other European country. While B3a, B9, (and an important sub-lineage of A2a5, namely, A2a5c) most likely originated in Spain, the other three haplogroups were imported from other European locations. The B3a strain may have originated in the Basque Country from a B3 ancestor of uncertain geographic origin, whereas B9 likely emerged in Madrid. The time of the most recent common ancestor (TMRCA) of SARS-CoV-2 suggested that the first coronavirus entered the country around 11 February 2020, as estimated from the TMRCA of B3a, the first lineage detected in the country. Moreover, earlier claims that the D614G mutation is associated to higher transmissibility is not consistent with the very high prevalence of COVID-19 in Spain when compared to other countries with lower disease incidence but much higher frequency of this mutation (56.4% in Spain vs. 82.4% in rest of Europe). Instead, the data support a major role of genetic drift in modeling the micro-geographic stratification of virus strains across the country as well as the role of SARS-CoV-2 super-spreaders.
西班牙曾经是新型冠状病毒肺炎(COVID-19)全球大流行的中心之一。本研究我们分析了从GISAID数据库上获取超过4100个病毒基因组(包括超过2600个高质量的基因组),其中包含了在西班牙采样的1245个基因组(922个高质量基因组)。这项研究的主要目的是分析SARS-CoV-2病毒的基因组变异情况,并重建西班牙病毒的谱系地理和传播模式。谱系地理分析表明,从疫情爆发开始SARS-CoV-2在西班牙至少有34次独立传入事件,其中6个谱系毒株非常成功地在西班牙传播,也就是六个超级传播者,A2a4型(7.8%),A2a5型(38.4%),A2a10型(2.8%),B3a型(30.1%)和B9型(8.7%)毒株所造成,这些毒株数据占西班牙所有基因组的87.9%。西班牙流行的SARS-CoV-2基因组中有一个独特B型谱系(39.3%,主要是B3a+B9型)毒株,其发生频率远高于其它任何欧洲国家。研究推测B3a型,B9型(以及A2a5型的一个重要衍生谱系,即A2a5c型)毒株最有可能起源于西班牙,其它三个单倍型毒株可能是由从其他欧洲地区输入。B3a谱系毒株可能起源于巴斯克地区,由一个地理来源不确定的B3祖先毒株衍生而来,而B9谱系毒株则可能来源于马德里。基于估算西班牙国内检测到的第一个谱系B3a最近共同祖先(TMRCA)分化时间,推测出第一批病毒输入西班牙的时间大约在2020年2月11日。此外,对比其他疾病发病率较低但D614G突变发生率更高的国家(西班牙56.4%对比欧洲其他地区82.4%),早先的说法是D614G突变体与较高的传播能力相关,但这与西班牙COVID-19的高流行并不相符。相反,这些数据支持遗传漂变在模拟病毒株微地理尺度下分区的主要作用,以及SARS-CoV-2超级传播者在传播病毒中的作用。.
Keywords: Covid-19; Genomics; Phylogeny; Phylogeography; SARS-CoV-2.
Figures

Similar articles
-
Mapping genome variation of SARS-CoV-2 worldwide highlights the impact of COVID-19 super-spreaders.Genome Res. 2020 Oct;30(10):1434-1448. doi: 10.1101/gr.266221.120. Epub 2020 Sep 2. Genome Res. 2020. PMID: 32878977 Free PMC article.
-
Genomic Analysis of Early SARS-CoV-2 Variants Introduced in Mexico.J Virol. 2020 Aug 31;94(18):e01056-20. doi: 10.1128/JVI.01056-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32641486 Free PMC article.
-
A Founder Effect Led Early SARS-CoV-2 Transmission in Spain.J Virol. 2021 Jan 13;95(3):e01583-20. doi: 10.1128/JVI.01583-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33127745 Free PMC article.
-
[Source of the COVID-19 pandemic: ecology and genetics of coronaviruses (Betacoronavirus: Coronaviridae) SARS-CoV, SARS-CoV-2 (subgenus Sarbecovirus), and MERS-CoV (subgenus Merbecovirus).].Vopr Virusol. 2020;65(2):62-70. doi: 10.36233/0507-4088-2020-65-2-62-70. Vopr Virusol. 2020. PMID: 32515561 Review. Russian.
-
The outbreak of the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): A review of the current global status.J Infect Public Health. 2020 Nov;13(11):1601-1610. doi: 10.1016/j.jiph.2020.07.011. Epub 2020 Aug 4. J Infect Public Health. 2020. PMID: 32778421 Free PMC article.
Cited by
-
Case Report: Genomic Characteristics of the First Known Case of SARS-CoV-2 Imported From Spain to Sichuan, China.Front Med (Lausanne). 2021 Nov 30;8:783646. doi: 10.3389/fmed.2021.783646. eCollection 2021. Front Med (Lausanne). 2021. PMID: 34917639 Free PMC article.
-
A Global Mutational Profile of SARS-CoV-2: A Systematic Review and Meta-Analysis of 368,316 COVID-19 Patients.Life (Basel). 2021 Nov 11;11(11):1224. doi: 10.3390/life11111224. Life (Basel). 2021. PMID: 34833100 Free PMC article. Review.
-
The phylodynamics of SARS-CoV-2 during 2020 in Finland.Commun Med (Lond). 2022 Jun 10;2:65. doi: 10.1038/s43856-022-00130-7. eCollection 2022. Commun Med (Lond). 2022. PMID: 35698660 Free PMC article.
-
COVID-19 Pandemic Waves and 2024-2025 Winter Season in Relation to Angiotensin-Converting Enzyme Inhibitors, Angiotensin Receptor Blockers and Amantadine.Healthcare (Basel). 2025 May 27;13(11):1270. doi: 10.3390/healthcare13111270. Healthcare (Basel). 2025. PMID: 40508884 Free PMC article.
-
Genomic Epidemiology of SARS-CoV-2 in Madrid, Spain, during the First Wave of the Pandemic: Fast Spread and Early Dominance by D614G Variants.Microorganisms. 2021 Feb 22;9(2):454. doi: 10.3390/microorganisms9020454. Microorganisms. 2021. PMID: 33671631 Free PMC article.
References
-
- Conrad O, Bechtel B, Bock M, Dietrich H, Fischer E, Gerlitz L, et al System for automated geoscientific analyses (SAGA) v.2.1.4. Geoscientific Model Development. 2015;8(7):1991–2007. doi: 10.5194/gmd-8-1991-2015. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous
