IKKγ/NEMO Is Required to Confer Antimicrobial Innate Immune Responses in the Yellow Mealworm, Tenebrio Molitor
- PMID: 32937897
- PMCID: PMC7555931
- DOI: 10.3390/ijms21186734
IKKγ/NEMO Is Required to Confer Antimicrobial Innate Immune Responses in the Yellow Mealworm, Tenebrio Molitor
Abstract
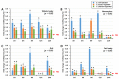
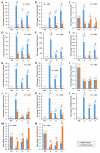
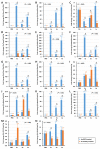
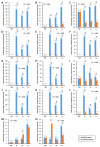
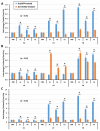
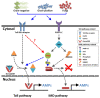
IKKγ/NEMO is the regulatory subunit of the IκB kinase (IKK) complex, which regulates the NF-κB signaling pathway. Within the IKK complex, IKKγ/NEMO is the non-catalytic subunit, whereas IKKα and IKKβ are the structurally related catalytic subunits. In this study, TmIKKγ was screened from the Tenebrio molitor RNA-Seq database and functionally characterized using RNAi screening for its role in regulating T. molitor antimicrobial peptide (AMP) genes after microbial challenges. The TmIKKγ transcript is 1521 bp that putatively encodes a polypeptide of 506 amino acid residues. TmIKKγ contains a NF-κB essential modulator (NEMO) and a leucine zipper domain of coiled coil region 2 (LZCC2). A phylogenetic analysis confirmed its homology to the red flour beetle, Tribolium castaneum IKKγ (TcIKKγ). The expression of TmIKKγ mRNA showed that it might function in diverse tissues of the insect, with a higher expression in the hemocytes and the fat body of the late-instar larvae. TmIKKγ mRNA expression was induced by Escherichia coli, Staphylococcus aureus, and Candida albicans challenges in the whole larvae and in tissues such as the hemocytes, gut and fat body. The knockdown of TmIKKγ mRNA significantly reduced the survival of the larvae after microbial challenges. Furthermore, we investigated the tissue-specific induction patterns of fourteen T. molitor AMP genes in TmIKKγ mRNA-silenced individuals after microbial challenges. In general, the mRNA expression of TmTenecin1, -2, and -4; TmDefensin1 and -2; TmColeoptericin1 and 2; and TmAttacin1a, 1b, and 2 were found to be downregulated in the hemocytes, gut, and fat body tissues in the TmIKKγ-silenced individuals after microbial challenges. Under similar conditions, TmRelish (NF-κB transcription factor) mRNA was also found to be downregulated. Thus, TmIKKγ is an important factor in the antimicrobial innate immune response of T. molitor.
Keywords: NF-κB transcription factor; RNAi; Tenebrio molitor; antimicrobial peptides; insect immunity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

