MAIRA- real-time taxonomic and functional analysis of long reads on a laptop
- PMID: 32938391
- PMCID: PMC7495841
- DOI: 10.1186/s12859-020-03684-2
MAIRA- real-time taxonomic and functional analysis of long reads on a laptop
Abstract
Background: Advances in mobile sequencing devices and laptop performance make metagenomic sequencing and analysis in the field a technologically feasible prospect. However, metagenomic analysis pipelines are usually designed to run on servers and in the cloud.
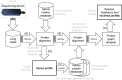
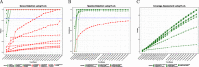
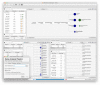
Results: MAIRA is a new standalone program for interactive taxonomic and functional analysis of long read metagenomic sequencing data on a laptop, without requiring external resources. The program performs fast, online, genus-level analysis, and on-demand, detailed taxonomic and functional analysis. It uses two levels of frame-shift-aware alignment of DNA reads against protein reference sequences, and then performs detailed analysis using a protein synteny graph.
Conclusions: We envision this software being used by researchers in the field, when access to servers or cloud facilities is difficult, or by individuals that do not routinely access such facilities, such as medical researchers, crop scientists, or teachers.
Keywords: Antibiotic resistance; Functional analysis; Long read sequencing; Metagenomics; Microbiome; Mobile computing; Open source software; Taxonomic analysis; Virulence factors.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Weber N, Liou D, Dommer J, MacMenamin P, Quinones M, Misner I, Oler AJ, Wan J, Kim L, Coakley McCarthy M, Ezeji S, Noble K, Hurt DE. Nephele: a cloud platform for simplified, standardized and reproducible microbiome data analysis. Bioinformatics. 2018;34(8):1411–3. doi: 10.1093/bioinformatics/btx617. - DOI - PMC - PubMed
-
- Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FSL, Wright GD, McArthur AG. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45(D1):566–73. doi: 10.1093/nar/gkw1004. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

