Multi-model seascape genomics identifies distinct environmental drivers of selection among sympatric marine species
- PMID: 32938400
- PMCID: PMC7493327
- DOI: 10.1186/s12862-020-01679-4
Multi-model seascape genomics identifies distinct environmental drivers of selection among sympatric marine species
Abstract
Background: As global change and anthropogenic pressures continue to increase, conservation and management increasingly needs to consider species' potential to adapt to novel environmental conditions. Therefore, it is imperative to characterise the main selective forces acting on ecosystems, and how these may influence the evolutionary potential of populations and species. Using a multi-model seascape genomics approach, we compare putative environmental drivers of selection in three sympatric southern African marine invertebrates with contrasting ecology and life histories: Cape urchin (Parechinus angulosus), Common shore crab (Cyclograpsus punctatus), and Granular limpet (Scutellastra granularis).
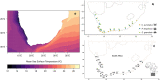
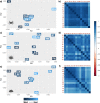
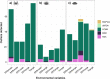
Results: Using pooled (Pool-seq), restriction-site associated DNA sequencing (RAD-seq), and seven outlier detection methods, we characterise genomic variation between populations along a strong biogeographical gradient. Of the three species, only S. granularis showed significant isolation-by-distance, and isolation-by-environment driven by sea surface temperatures (SST). In contrast, sea surface salinity (SSS) and range in air temperature correlated more strongly with genomic variation in C. punctatus and P. angulosus. Differences were also found in genomic structuring between the three species, with outlier loci contributing to two clusters in the East and West Coasts for S. granularis and P. angulosus, but not for C. punctatus.
Conclusion: The findings illustrate distinct evolutionary potential across species, suggesting that species-specific habitat requirements and responses to environmental stresses may be better predictors of evolutionary patterns than the strong environmental gradients within the region. We also found large discrepancies between outlier detection methodologies, and thus offer a novel multi-model approach to identifying the principal environmental selection forces acting on species. Overall, this work highlights how adding a comparative approach to seascape genomics (both with multiple models and species) can elucidate the intricate evolutionary responses of ecosystems to global change.
Keywords: Comparative phylogeography; Environmental association; Marine invertebrates; Pool-seq; RAD-seq; Seascape genomics.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Pecl GT, Ward TM, Doubleday ZA, Clarke S, Day J, Dixon C, et al. Rapid assessment of fisheries species sensitivity to climate change. Clim Chang. 2014;127(3–4):505–520.
-
- Glavovic B, Limburg K, Liu K-K, Emeis K-C, Thomas H, Kremer H, et al. Living on the margin in the Anthropocene: engagement arenas for sustainability research and action at the ocean–land interface. Curr Opin Environ Sust. 2015;14:232–238.
-
- Helmuth B, Mieszkowska N, Moore P, Hawkins SJ. Living on the edge of two changing worlds: forecasting the responses of rocky intertidal ecosystems to climate change. Annu Rev Ecol Evol Syst. 2006;37(1):373–404.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

