Microbial dark matter filling the niche in hypersaline microbial mats
- PMID: 32938503
- PMCID: PMC7495880
- DOI: 10.1186/s40168-020-00910-0
Microbial dark matter filling the niche in hypersaline microbial mats
Abstract
Background: Shark Bay, Australia, harbours one of the most extensive and diverse systems of living microbial mats that are proposed to be analogs of some of the earliest ecosystems on Earth. These ecosystems have been shown to possess a substantial abundance of uncultivable microorganisms. These enigmatic microbes, jointly coined as 'microbial dark matter' (MDM), are hypothesised to play key roles in modern microbial mats.
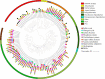
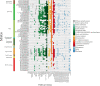
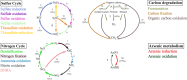
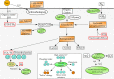
Results: We reconstructed 115 metagenome-assembled genomes (MAGs) affiliated to MDM, spanning 42 phyla. This study reports for the first time novel microorganisms (Zixibacterial order GN15) putatively taking part in dissimilatory sulfate reduction in surface hypersaline settings, as well as novel eukaryote signature proteins in the Asgard archaea. Despite possessing reduced-size genomes, the MDM MAGs are capable of fermenting and degrading organic carbon, suggesting a role in recycling organic carbon. Several forms of RuBisCo were identified, allowing putative CO2 incorporation into nucleotide salvaging pathways, which may act as an alternative carbon and phosphorus source. High capacity of hydrogen production was found among Shark Bay MDM. Putative schizorhodopsins were also identified in Parcubacteria, Asgard archaea, DPANN archaea, and Bathyarchaeota, allowing these members to potentially capture light energy. Diversity-generating retroelements were prominent in DPANN archaea that likely facilitate the adaptation to a dynamic, host-dependent lifestyle.
Conclusions: This is the first study to reconstruct and describe in detail metagenome-assembled genomes (MAGs) affiliated with microbial dark matter in hypersaline microbial mats. Our data suggests that these microbial groups are major players in these systems. In light of our findings, we propose H2, ribose and CO/CO2 as the main energy currencies of the MDM community in these mat systems. Video Abstract.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Unravelling core microbial metabolisms in the hypersaline microbial mats of Shark Bay using high-throughput metagenomics.ISME J. 2016 Jan;10(1):183-96. doi: 10.1038/ismej.2015.87. Epub 2015 May 29. ISME J. 2016. PMID: 26023869 Free PMC article.
-
Genome-resolved metagenomics provides insights into the functional complexity of microbial mats in Blue Holes, Shark Bay.FEMS Microbiol Ecol. 2022 Jan 19;98(1):fiab158. doi: 10.1093/femsec/fiab158. FEMS Microbiol Ecol. 2022. PMID: 34865013
-
Disentangling the drivers of functional complexity at the metagenomic level in Shark Bay microbial mat microbiomes.ISME J. 2018 Nov;12(11):2619-2639. doi: 10.1038/s41396-018-0208-8. Epub 2018 Jul 6. ISME J. 2018. PMID: 29980796 Free PMC article.
-
Major New Microbial Groups Expand Diversity and Alter our Understanding of the Tree of Life.Cell. 2018 Mar 8;172(6):1181-1197. doi: 10.1016/j.cell.2018.02.016. Cell. 2018. PMID: 29522741 Review.
-
Molecular Ecology of Hypersaline Microbial Mats: Current Insights and New Directions.Microorganisms. 2016 Jan 5;4(1):6. doi: 10.3390/microorganisms4010006. Microorganisms. 2016. PMID: 27681900 Free PMC article. Review.
Cited by
-
Defining Composition and Function of the Rhizosphere Microbiota of Barley Genotypes Exposed to Growth-Limiting Nitrogen Supplies.mSystems. 2022 Dec 20;7(6):e0093422. doi: 10.1128/msystems.00934-22. Epub 2022 Nov 7. mSystems. 2022. PMID: 36342125 Free PMC article.
-
Phylogenetically and metabolically diverse autotrophs in the world's deepest blue hole.ISME Commun. 2023 Nov 14;3(1):117. doi: 10.1038/s43705-023-00327-4. ISME Commun. 2023. PMID: 37964026 Free PMC article.
-
The Evolutionary Kaleidoscope of Rhodopsins.mSystems. 2022 Oct 26;7(5):e0040522. doi: 10.1128/msystems.00405-22. Epub 2022 Sep 19. mSystems. 2022. PMID: 36121162 Free PMC article.
-
Community ecology and functional potential of bacteria, archaea, eukarya and viruses in Guerrero Negro microbial mat.Sci Rep. 2024 Jan 31;14(1):2561. doi: 10.1038/s41598-024-52626-y. Sci Rep. 2024. PMID: 38297006 Free PMC article.
-
The ecological roles of microbial lipopeptides: Where are we going?Comput Struct Biotechnol J. 2021 Mar 2;19:1400-1413. doi: 10.1016/j.csbj.2021.02.017. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 33777336 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

