Pharmacologic Suppression of B7-H4 Glycosylation Restores Antitumor Immunity in Immune-Cold Breast Cancers
- PMID: 32938586
- PMCID: PMC7710601
- DOI: 10.1158/2159-8290.CD-20-0402
Pharmacologic Suppression of B7-H4 Glycosylation Restores Antitumor Immunity in Immune-Cold Breast Cancers
Abstract
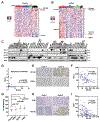
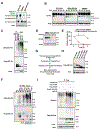
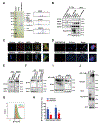
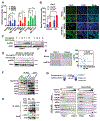
Despite widespread utilization of immunotherapy, treating immune-cold tumors has proved to be a challenge. Here, we report that expression of the immune checkpoint molecule B7-H4 is prevalent among immune-cold triple-negative breast cancers (TNBC), where its expression inversely correlates with that of PD-L1. Glycosylation of B7-H4 interferes with its interaction/ubiquitination by AMFR, resulting in B7-H4 stabilization. B7-H4 expression inhibits doxorubicin-induced cell death through the suppression of eIF2α phosphorylation required for calreticulin exposure vis-à-vis the cancer cells. NGI-1, which inhibits B7-H4 glycosylation causing its ubiquitination and subsequent degradation, improves the immunogenic properties of cancer cells treated with doxorubicin, enhancing their phagocytosis by dendritic cells and their capacity to elicit CD8+ IFNγ-producing T-cell responses. In preclinical models of TNBC, a triple combination of NGI-1, camsirubicin (a noncardiotoxic doxorubicin analogue) and PD-L1 blockade was effective in reducing tumor growth. Collectively, our findings uncover a strategy for targeting the immunosuppressive molecule B7-H4. SIGNIFICANCE: This work unravels the regulation of B7-H4 stability by ubiquitination and glycosylation, which affects tumor immunogenicity, particularly regarding immune-cold breast cancers. The inhibition of B7-H4 glycosylation can be favorably combined with immunogenic chemotherapy and PD-L1 blockade to achieve superior immuno-infiltration of cold tumors, as well as improved tumor growth control.See related commentary by Pearce and Läubli, p. 1789.This article is highlighted in the In This Issue feature, p. 1775.
©2020 American Association for Cancer Research.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest
The authors declare no potential conflicts of interest.
Figures



Comment in
-
A Sweet Approach to Heat Up Cancer Response to Immunotherapy.Cancer Discov. 2020 Dec;10(12):1789-1790. doi: 10.1158/2159-8290.CD-20-1355. Cancer Discov. 2020. PMID: 33262181
References
-
- Sica GL, Choi IH, Zhu G, Tamada K, Wang SD, Tamura H, et al. B7-H4, a molecule of the B7 family, negatively regulates T cell immunity. Immunity 2003;18(6):849–61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

