Translational induction of ATF4 during integrated stress response requires noncanonical initiation factors eIF2D and DENR
- PMID: 32938929
- PMCID: PMC7495428
- DOI: 10.1038/s41467-020-18453-1
Translational induction of ATF4 during integrated stress response requires noncanonical initiation factors eIF2D and DENR
Abstract
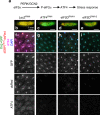
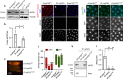
The Integrated Stress Response (ISR) helps metazoan cells adapt to cellular stress by limiting the availability of initiator methionyl-tRNA for translation. Such limiting conditions paradoxically stimulate the translation of ATF4 mRNA through a regulatory 5' leader sequence with multiple upstream Open Reading Frames (uORFs), thereby activating stress-responsive gene expression. Here, we report the identification of two critical regulators of such ATF4 induction, the noncanonical initiation factors eIF2D and DENR. Loss of eIF2D and DENR in Drosophila results in increased vulnerability to amino acid deprivation, susceptibility to retinal degeneration caused by endoplasmic reticulum (ER) stress, and developmental defects similar to ATF4 mutants. eIF2D requires its RNA-binding motif for regulation of 5' leader-mediated ATF4 translation. Consistently, eIF2D and DENR deficient human cells show impaired ATF4 protein induction in response to ER stress. Altogether, our findings indicate that eIF2D and DENR are critical mediators of ATF4 translational induction and stress responses in vivo.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Delépine M, et al. EIF2AK3, encoding translation initiation factor 2-alpha kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat. Genet. 2000;25:406–409. - PubMed
-
- Harding HP, et al. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol. Cell. 2001;7:1153–1163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

