Benchmarking microbial growth rate predictions from metagenomes
- PMID: 32939027
- PMCID: PMC7852909
- DOI: 10.1038/s41396-020-00773-1
Benchmarking microbial growth rate predictions from metagenomes
Abstract
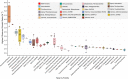
Growth rates are central to understanding microbial interactions and community dynamics. Metagenomic growth estimators have been developed, specifically codon usage bias (CUB) for maximum growth rates and "peak-to-trough ratio" (PTR) for in situ rates. Both were originally tested with pure cultures, but natural populations are more heterogeneous, especially in individual cell histories pertinent to PTR. To test these methods, we compared predictors with observed growth rates of freshly collected marine prokaryotes in unamended seawater. We prefiltered and diluted samples to remove grazers and greatly reduce virus infection, so net growth approximated gross growth. We sampled over 44 h for abundances and metagenomes, generating 101 metagenome-assembled genomes (MAGs), including Actinobacteria, Verrucomicrobia, SAR406, MGII archaea, etc. We tracked each MAG population by cell-abundance-normalized read recruitment, finding growth rates of 0 to 5.99 per day, the first reported rates for several groups, and used these rates as benchmarks. PTR, calculated by three methods, rarely correlated to growth (r ~-0.26-0.08), except for rapidly growing γ-Proteobacteria (r ~0.63-0.92), while CUB correlated moderately well to observed maximum growth rates (r = 0.57). This suggests that current PTR approaches poorly predict actual growth of most marine bacterial populations, but maximum growth rates can be approximated from genomic characteristics.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Kirchman DL. Growth rates of microbes in the oceans. Annu Rev Mar Sci. 2016;8:285–309. - PubMed
-
- Koch BJ, McHough TA, Hayer M, Schwartz E, Blazewicz SJ, Dijkstra P, et al. Estimating taxon-specific population dynamics in diverse microbial communities. Ecosphere. 2018;9:e02090.
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

