Modelling kidney disease using ontology: insights from the Kidney Precision Medicine Project
- PMID: 32939051
- PMCID: PMC8012202
- DOI: 10.1038/s41581-020-00335-w
Modelling kidney disease using ontology: insights from the Kidney Precision Medicine Project
Abstract
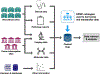
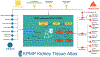
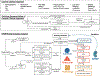
An important need exists to better understand and stratify kidney disease according to its underlying pathophysiology in order to develop more precise and effective therapeutic agents. National collaborative efforts such as the Kidney Precision Medicine Project are working towards this goal through the collection and integration of large, disparate clinical, biological and imaging data from patients with kidney disease. Ontologies are powerful tools that facilitate these efforts by enabling researchers to organize and make sense of different data elements and the relationships between them. Ontologies are critical to support the types of big data analysis necessary for kidney precision medicine, where heterogeneous clinical, imaging and biopsy data from diverse sources must be combined to define a patient's phenotype. The development of two new ontologies - the Kidney Tissue Atlas Ontology and the Ontology of Precision Medicine and Investigation - will support the creation of the Kidney Tissue Atlas, which aims to provide a comprehensive molecular, cellular and anatomical map of the kidney. These ontologies will improve the annotation of kidney-relevant data, and eventually lead to new definitions of kidney disease in support of precision medicine.
Conflict of interest statement
Competing interests
The authors declare no conflicts of interest.
Figures
References
-
- Khwaja A KDIGO clinical practice guidelines for acute kidney injury. Nephron Clin. Pract 120, c179–84 (2012). - PubMed
-
- Stevens PE, Levin A & Kidney Disease: Improving Global Outcomes Chronic Kidney Disease Guideline Development Work Group Members. Evaluation and management of chronic kidney disease: synopsis of the kidney disease: improving global outcomes 2012 clinical practice guideline. Ann. Intern. Med 158, 825–830 (2013). - PubMed
-
- Hawrylycz M et al. The Allen Brain Atlas. Springer Handbook of Bio-/Neuroinformatics 1111–1126 (2014) doi:10.1007/978-3-642-30574-0_62. - DOI

